 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG004152.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 318aa MW: 35529.6 Da PI: 6.374 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.2 | 1.7e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd +lv +++++G+g+W++++ g+ R+ k+c++rw +yl
CCG004152.1 14 KGPWTPEEDIILVSYIQEHGPGNWRAVPTNTGLLRCSKSCRLRWTNYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48.6 | 1.8e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T E++ ++++ ++lG++ W+tIa++++ Rt++++k++w+++l
CCG004152.1 67 RGNFTHSEEKMIIHLQALLGNR-WATIASYLP-QRTDNDIKNYWNTHL 112
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-23 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.649 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.87E-30 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.2E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.20E-11 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 23.485 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-23 | 65 | 114 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-14 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.2E-14 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.47E-10 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001666 | Biological Process | response to hypoxia | ||||
| GO:0009617 | Biological Process | response to bacterium | ||||
| GO:0009626 | Biological Process | plant-type hypersensitive response | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0042761 | Biological Process | very long-chain fatty acid biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 318 aa Download sequence Send to blast |
MGRPPCCDKT GVMKGPWTPE EDIILVSYIQ EHGPGNWRAV PTNTGLLRCS KSCRLRWTNY 60 LRPGIKRGNF THSEEKMIIH LQALLGNRWA TIASYLPQRT DNDIKNYWNT HLKKLKKLQA 120 GQEGQSRDGI SSTGSQQISR GQWERRLQTD IHMARQALSE ALSPEKPNSL LTELKPSSGY 180 EKPAPASLYA SSTENIAKLL KGWMRSGPNQ SLTNSTTPQN SFNNIAVADS FSSEETINRA 240 DENGTELSEA FESLFGFDSS NINFSRSSPD TGLLQDESKP NSSAQVPLSV IERWLFDEGA 300 MEGKEYLSEV TPDENNLF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 7e-24 | 12 | 114 | 56 | 157 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 7e-24 | 12 | 114 | 56 | 157 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 6e-24 | 12 | 114 | 2 | 103 | C-Myb DNA-Binding Domain |
| 1msf_C | 6e-24 | 12 | 114 | 2 | 103 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00386 | DAP | Transfer from AT3G28910 | Download |
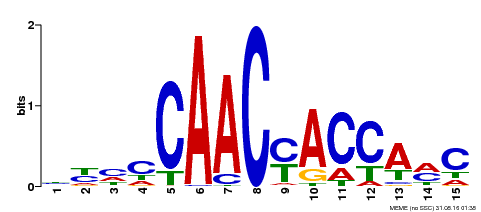
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC216459 | 0.0 | AC216459.1 Populus trichocarpa clone POP074-L11, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011013219.1 | 0.0 | PREDICTED: myb-related protein 306-like | ||||
| Swissprot | P81392 | 1e-112 | MYB06_ANTMA; Myb-related protein 306 | ||||
| TrEMBL | B9H0F8 | 0.0 | B9H0F8_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0004s12530.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1016 | 34 | 115 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G28910.1 | 1e-108 | myb domain protein 30 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



