 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG013474.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 201aa MW: 21102.8 Da PI: 9.337 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.4 | 7.4e-39 | 32 | 92 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++++l+cprCdstntkfCyynny+ sqPr+fCk+CrryWt+GG+lr++PvGgg+rkn k+s
CCG013474.1 32 EQEHLPCPRCDSTNTKFCYYNNYNFSQPRHFCKSCRRYWTHGGTLRDIPVGGGTRKNAKRS 92
68899****************************************************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 5.0E-29 | 29 | 90 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 2.1E-33 | 34 | 90 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.255 | 36 | 90 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 38 | 74 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 201 aa Download sequence Send to blast |
MPAELSSSEA ARRPQQSTAA TLTKPGGAPP QEQEHLPCPR CDSTNTKFCY YNNYNFSQPR 60 HFCKSCRRYW THGGTLRDIP VGGGTRKNAK RSRTSATSPA SFVGPITGTN IDGLPLPATP 120 VLLPLTSNQG LSVHFGGGDG KGNGGGLGGS FTSLLNTQGP AGFLALNGFG LGIGPGIEDV 180 NFALGRGLWP FPGMWEMLAL P |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00593 | DAP | Transfer from AT5G66940 | Download |
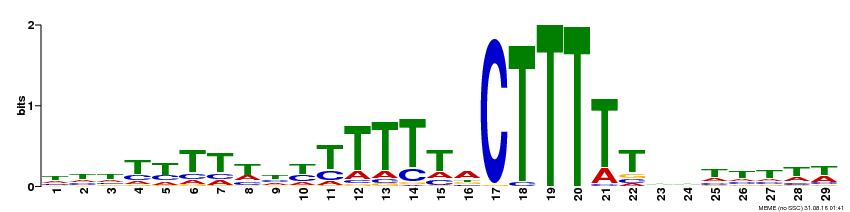
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC212845 | 0.0 | AC212845.1 Populus trichocarpa clone POP021-P21, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011039246.1 | 1e-139 | PREDICTED: LOW QUALITY PROTEIN: dof zinc finger protein DOF3.4 | ||||
| Swissprot | Q9FGD6 | 2e-52 | DOF58_ARATH; Dof zinc finger protein DOF5.8 | ||||
| TrEMBL | B9HEE8 | 1e-122 | B9HEE8_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0007s11790.1 | 1e-123 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2801 | 31 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G50410.1 | 2e-41 | OBF binding protein 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



