 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG018924.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 238aa MW: 27211.3 Da PI: 4.6722 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61.8 | 1e-19 | 30 | 84 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++++f++eq++ Le++Fe++ + +++ +LAk+lgL+ rqV +WFqN+Ra++k
CCG018924.1 30 KNKRRFSDEQIKSLESMFESETRLEPRKKMQLAKELGLQPRQVAIWFQNKRARWK 84
67889*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 121.3 | 4.8e-39 | 31 | 122 | 2 | 93 |
HD-ZIP_I/II 2 kkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
+krr+s+eq+k+LE++Fe+e++Lep++K++la+eLglqprqva+WFqn+RAR+k+kqlE+d++ L+++y++l+++ e+L+ke+++L +l++
CCG018924.1 31 NKRRFSDEQIKSLESMFESETRLEPRKKMQLAKELGLQPRQVAIWFQNKRARWKSKQLERDFSILRANYNSLASRFETLKKEKQALVLQLQK 122
79************************************************************************************999876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.97E-20 | 8 | 88 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-20 | 20 | 87 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.544 | 26 | 86 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.9E-17 | 28 | 90 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 5.0E-17 | 30 | 84 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.34E-15 | 35 | 87 | No hit | No description |
| PRINTS | PR00031 | 6.2E-6 | 57 | 66 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 61 | 84 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 6.2E-6 | 66 | 82 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 5.1E-15 | 86 | 127 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 238 aa Download sequence Send to blast |
MFDGGEYSPS ATEPFSCMNG VTTSRKKKNK NKRRFSDEQI KSLESMFESE TRLEPRKKMQ 60 LAKELGLQPR QVAIWFQNKR ARWKSKQLER DFSILRANYN SLASRFETLK KEKQALVLQL 120 QKINGLMKKP GEEGECCDQG PAVNSIEGRS ENADTTMGES ETNPRLSIER PEHGLGVLSD 180 EDSSIKAEYF ELEEEPNLIS MVEPADGSLT SQEDWGSLDS DGLFDQSSSD YQWWDFWA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription activator that may act as growth regulators in response to water deficit. {ECO:0000269|PubMed:15604708, ECO:0000269|PubMed:8771791}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00650 | PBM | Transfer from LOC_Os09g35910 | Download |
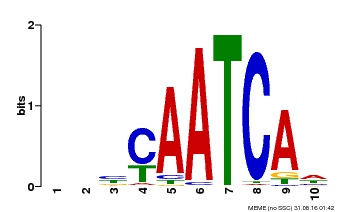
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By water deficit, by abscisic acid (ABA) and by salt stress. {ECO:0000269|PubMed:16055682, ECO:0000269|PubMed:8771791}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CU226615 | 0.0 | CU226615.1 Populus EST from severe drought-stressed leaves. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011023121.1 | 1e-176 | PREDICTED: homeobox-leucine zipper protein ATHB-7-like | ||||
| Swissprot | P46897 | 1e-75 | ATHB7_ARATH; Homeobox-leucine zipper protein ATHB-7 | ||||
| TrEMBL | B9GR61 | 1e-161 | B9GR61_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0002s17680.1 | 1e-162 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5635 | 34 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46680.1 | 1e-73 | homeobox 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



