 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG019165.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1002aa MW: 110956 Da PI: 6.4263 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 132.6 | 1.3e-41 | 150 | 227 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C +dls+ak+yhrrhkvCe+hska+++lv +++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
CCG019165.1 150 VCQVEDCGVDLSNAKDYHRRHKVCEMHSKASKALVGNVMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 227
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.6E-33 | 146 | 212 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.308 | 148 | 225 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.62E-39 | 149 | 230 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.7E-30 | 151 | 224 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 6.1E-7 | 302 | 356 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.51E-7 | 776 | 886 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.1E-7 | 779 | 887 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.66E-7 | 779 | 886 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1002 aa Download sequence Send to blast |
MEARFGGEAH HFYAMGPTDM RAVGKRGLEW DLNDWKWDGD LFIASPLNPV PSTSVRRPFF 60 PLGVGTGVPA TGNSTNSSSS CSDEVNLGVE KGKRELEKRR RVVVIEDDNL NDQETGGLSL 120 KLGGQRDAGN WEGSSGKKTK LVGGGLSRAV CQVEDCGVDL SNAKDYHRRH KVCEMHSKAS 180 KALVGNVMQR FCQQCSRFHV LQEFDEGKRS CRRRLAGHNK RRRKTNPDTV GNGSSMNDDQ 240 NSGYLLISLL RILSNMHSNR SDETTDQDLL THLLRSLATH SVEHGGRNMF GPLQEPRDLS 300 TSFGNSEVVS TLLSNGEGPS NLKQPLTVPV SGMPQQVVPV HDAYGANIQT TSSLKPSIPN 360 NFAVYSEVRE STAGQVKMNN FDLNDIYVDS DDGAEDIERS PAPVNARTSS LDCPSWVQQD 420 SHQSSPPQTS RNSDSASAQS PSSSSGEAQS RTDRIVFKLF GKEPNDFPLV LRAQILDWLS 480 HSPTDIESYI RPGCIILTIY LHQAEAAWEE LCCGLGSSLS RLLDVSDDTF WRTGWIYIRV 540 QHQIAFVYNG QVVVDTSLPL TSNNYSKILS VKPIAITASE RAEFLIKGVN LSRPATRLLC 600 AVEGNYMVQE NTQEVMDGVD SFKGHDEVQC VNFSCSIPMV TGRGFIEIED HGFSSSFFPF 660 LVAEEDVCSE IRMLEGVLET ETDADFEETE KMAAKNQAMN FVHEMSWLLH RSQLKSRLGC 720 SDPSMNLFPL RRFKWLMEFS MDHEWCAVVG KLLSILHNGI VGTEEHSSLN VALSEMGLLH 780 RAVRRNSRSL VELLLRYVPE KFGSKDKALV GGSHESILFR PDVTGPAGLT PLHIAAGKDG 840 SEDVLDILTE DPGMVGIEAW KNALDSTGFT PEDYARLRGH YTYIHLVQRK INKRQAVGGH 900 VVLDIPSNLS NSNINVKQNE GLSSSFEIGR TALRPTQRNC KLCSQKVVYG IASRSQLYRP 960 AMLSMVAIAA VCVCVALLFK SCPEVLYVFR PFRWEMLDYG TS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-30 | 150 | 224 | 10 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 97 | 101 | KRRRV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
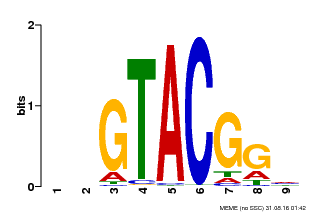
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011005939.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 isoform X1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | B9GSZ5 | 0.0 | B9GSZ5_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0002s18970.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1539 | 34 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



