 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG019351.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 378aa MW: 42454.1 Da PI: 9.2396 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.7 | 9.9e-32 | 63 | 116 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH +Fv+ave+LGG+e+AtPk +l+lm+v+gL+++hvkSHLQ+YR+
CCG019351.1 63 PRLRWTPELHLCFVKAVERLGGQERATPKLVLQLMNVNGLSIAHVKSHLQMYRS 116
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.975 | 59 | 119 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-29 | 59 | 117 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 7.35E-15 | 62 | 117 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.3E-23 | 63 | 118 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.5E-10 | 64 | 115 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MEESNHTGCS KTSASIQDHD ESAGGENDEE ESRPKKGGSS SNSTVEESEN KSSVRPYVRS 60 KLPRLRWTPE LHLCFVKAVE RLGGQERATP KLVLQLMNVN GLSIAHVKSH LQMYRSKKVD 120 DPSQGMADHR HLMESGDRNI YNLSQLPMLQ GYNQYQRQNS SFRYGDASWN AREHFIYNPH 180 VGRCVIDRTR PGSYGTVAER IFGSSNNSNW SANSAKFQMG ASSLISQSKW KNEELKGDQQ 240 LPQSLHNNRF WQPQPSPSLI DVSPLVLPQM QTKVGESSTH FKRFLPSDSK SATSTVQEWK 300 TLKRKASDCN LDLDLSLKLT PTKDHDSNQQ SLEDSTKVNS ELSLSLYSPS SSKLSRLKRE 360 GDGNKDHGKR SSTLDLTI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 4e-18 | 64 | 118 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-18 | 64 | 118 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-18 | 64 | 118 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-18 | 64 | 118 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 5e-18 | 64 | 118 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
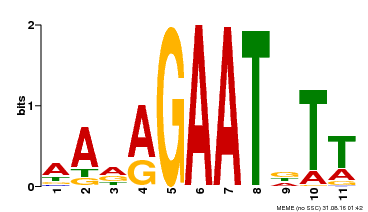
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC216544 | 0.0 | AC216544.1 Populus trichocarpa clone POP016-D10, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011046839.1 | 0.0 | PREDICTED: putative Myb family transcription factor At1g14600 | ||||
| Refseq | XP_011046840.1 | 0.0 | PREDICTED: putative Myb family transcription factor At1g14600 | ||||
| TrEMBL | A0A2K1XEP6 | 0.0 | A0A2K1XEP6_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0016s12940.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2378 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 3e-56 | G2-like family protein | ||||



