 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG019932.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 321aa MW: 35947.9 Da PI: 6.7953 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59.1 | 9.6e-19 | 29 | 76 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd +l+++++ +G g+W++ ar+ g++Rt+k+c++rw++yl
CCG019932.1 29 RGPWTVEEDFKLINYIATHGEGRWNSLARCAGLKRTGKSCRLRWLNYL 76
89********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 49.5 | 9.9e-16 | 82 | 125 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ T eE++ +++++ ++G++ W++Ia++++ gRt++++k++w++
CCG019932.1 82 RGNITLEEQLMILELHSRWGNR-WSKIAQHLP-GRTDNEIKNYWRT 125
7999******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.072 | 24 | 76 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.67E-31 | 27 | 123 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.9E-16 | 28 | 78 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.9E-17 | 29 | 76 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-23 | 30 | 83 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.52E-13 | 31 | 76 | No hit | No description |
| PROSITE profile | PS51294 | 22.707 | 77 | 131 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.6E-14 | 81 | 129 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-14 | 82 | 125 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.8E-23 | 84 | 130 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.29E-11 | 86 | 125 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009620 | Biological Process | response to fungus | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 321 aa Download sequence Send to blast |
MSMMDVKGNS NSSSSTTQSD EEMMGDLRRG PWTVEEDFKL INYIATHGEG RWNSLARCAG 60 LKRTGKSCRL RWLNYLRPDV RRGNITLEEQ LMILELHSRW GNRWSKIAQH LPGRTDNEIK 120 NYWRTRVQKH AKQLKCDVNS KQFKDTMRYL WMPRLIERIQ AAAAATATNT SSVSTAASVS 180 PAATVSAPAN NQHSISNSDV GTGNLVMAPH GINGNDFGVS HVTQSYTPEN SSTAASSDSF 240 ATQVSPVSDL TTDYYNIPIN RNTNPDYFQA GQVGYLESMI SPPGIFNHGL DFQAMEHSNN 300 QWPMDGGDTS DNMWNLEDIW F |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 3e-25 | 26 | 123 | 24 | 120 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor contributing to the regulation of stamen maturation and male fertility in response to jasmonate signaling. Required for correct timing of anther dehiscence. Acts as a negative regulator of abscisic acid-induced cell death. Not involved in the regulation of BOI. Regulated by MYB21 and at a lower level by MYB24. Negatively regulated by the proteasome in an SCF(COI1) E3 ubiquitin-protein ligase complex-dependent manner. {ECO:0000269|PubMed:14555693, ECO:0000269|PubMed:19091873, ECO:0000269|PubMed:20921156, ECO:0000269|PubMed:23952703}. | |||||
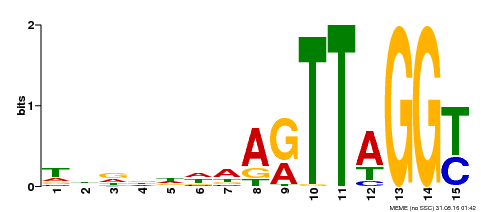
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00334 | DAP | Transfer from AT3G06490 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by jasmonate and pathogen infection. {ECO:0000269|PubMed:14555693, ECO:0000269|PubMed:19091873}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011016230.1 | 0.0 | PREDICTED: transcription factor MYB108-like | ||||
| Swissprot | Q9LDE1 | 3e-96 | MY108_ARATH; Transcription factor MYB108 | ||||
| TrEMBL | B9HIK1 | 0.0 | B9HIK1_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0008s10080.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF875 | 34 | 125 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G06490.1 | 5e-91 | myb domain protein 108 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



