 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG020009.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 340aa MW: 37742.6 Da PI: 6.8832 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 61.6 | 1.6e-19 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++lv++++++G g+W++ ++ g++R++k+c++rw +yl
CCG020009.1 14 KGPWTPEEDQKLVKYIQKHGHGSWRALPKLSGLNRCGKSCRLRWTNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 54.9 | 2e-17 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++++eE++ +++++ lG++ W++Ia++++ gRt++++k++w+++l
CCG020009.1 67 RGKFSQEEEQTILNLHSVLGNK-WSAIASHLP-GRTDNEIKNFWNTHL 112
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-26 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.138 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.35E-32 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.9E-16 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.9E-18 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.05E-13 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 26.627 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.7E-28 | 65 | 117 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.8E-17 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.2E-16 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.52E-12 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 340 aa Download sequence Send to blast |
MGRSPCCDES GLKKGPWTPE EDQKLVKYIQ KHGHGSWRAL PKLSGLNRCG KSCRLRWTNY 60 LRPDIKRGKF SQEEEQTILN LHSVLGNKWS AIASHLPGRT DNEIKNFWNT HLKKKLIQMG 120 FDPMTHRPRT DIFSSLPHLI ALANLKELID HHSLEEHALR LQTETAQMAK LQYLQYLLQP 180 QPPAASNTAT SSNNLNNIGT LSDIEAFNLL NSLASLKDSP VSSLSQLDLP ASSLQGINDS 240 IHFSHLPDLQ IPCNYQTPPT KGMAQAPELT VFSQGENSPI SPFQFPSSST PFPPSAVPPV 300 TQISSMVNNL GDASSTSSYG EEVPSVWHDL LEDPLFHDIA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-31 | 12 | 116 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250|UniProtKB:Q9M2D9}. | |||||
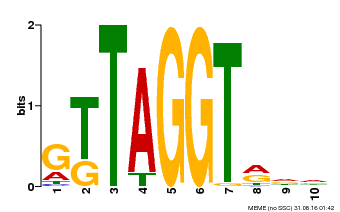
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00503 | DAP | Transfer from AT5G10280 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by gravity in roots. {ECO:0000269|PubMed:24902892}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011022802.1 | 0.0 | PREDICTED: myb-related protein 308-like | ||||
| Swissprot | Q9SBF3 | 1e-108 | MYB92_ARATH; Transcription factor MYB92 | ||||
| TrEMBL | A0A2K1ZRS4 | 0.0 | A0A2K1ZRS4_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0007s05360.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF402 | 34 | 172 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34670.1 | 2e-99 | myb domain protein 93 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



