 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG021402.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 779aa MW: 90432.6 Da PI: 8.6556 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 83.4 | 3.6e-26 | 98 | 207 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk..................tekerrtraetrtgCkaklkvkkek.dgkwevt 79
+fY+eYAk+vGFs+ +++s++s+ +g++++++fvC+++g+++++++ +++ r +++ ++t+Cka+++vk+++ dg+w+v
CCG021402.1 98 SFYKEYAKSVGFSTITKASRRSRISGKFIDAKFVCTRYGTKRDTSTIelpqpvsnadaatslpvkRKRGRINQSWSKTDCKACMHVKRRQqDGRWVVR 195
5***************************************99988879**************998888899******************99******* PP
FAR1 80 kleleHnHelap 91
++ +eHnHe++p
CCG021402.1 196 SFIKEHNHEIFP 207
*********975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 7.1E-24 | 98 | 207 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 2.9E-33 | 326 | 418 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 10.311 | 605 | 642 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 9.2E-7 | 617 | 644 | IPR006564 | Zinc finger, PMZ-type |
| Pfam | PF04434 | 1.0E-6 | 618 | 640 | IPR007527 | Zinc finger, SWIM-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 779 aa Download sequence Send to blast |
MGIDLEQPSG EYHKEDRRPN VNVNTVDGGD GGHERDQIIV NSPDIGGNGC EKTGTVINGR 60 VLDGRKKPNA GDRINLNSVK DAEPHDGMEF ESKDEAFSFY KEYAKSVGFS TITKASRRSR 120 ISGKFIDAKF VCTRYGTKRD TSTIELPQPV SNADAATSLP VKRKRGRINQ SWSKTDCKAC 180 MHVKRRQQDG RWVVRSFIKE HNHEIFPDQA YYFRCHRNLN LGNDNVDALH AIRARTKKLY 240 VAMSRQSGGH RKHENQKGGV TNPSGNTKHL ALDEGDAQAM LDHFIHMQDE NPNFFYAIDL 300 NEEQQLRNVF WVDAKGRLDY GNFGDVIFFD TTYLKNEYKL PFAPFIGVNH HFQFLLLGCA 360 LVADETKTTY VWLMRAWLRA MGGHAPRVIL TDQDTALKEA IQEVFPNSRH CFCLWHVFSK 420 IPEKLSYVTR QHENFMLKFK KCIFKSWTSE QFEKKWWKMV EIFNLRNDVW FQSLYEDRQR 480 WIPFFMRDNF LAGMSTTQRS ESINTLFDRY MQRKTTLREF LEQQKAMLQE KFEEEAKADF 540 ETWHKQPGLK SPSPFGKQMA SIYTHAIFKK FQVEVLGVVA CHPRKETEDG ETQTFKVQDF 600 EDNQYFIVVW NEMTSYLSCS CRLFEFNGFL CRHVLIVMQM SGLHSIPSQY ILKRWTKDAK 660 SRQIMREQSD VVESRVQRYN DLCRRAFKLG DEGSLSQESY NIAFNALEEA LRKCESVNNL 720 IQNIIEPTSP PSNGPLDYDE VNQAHGATKT NKKKDTSRKK QVSWVFGVCL LEVLMIFYV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
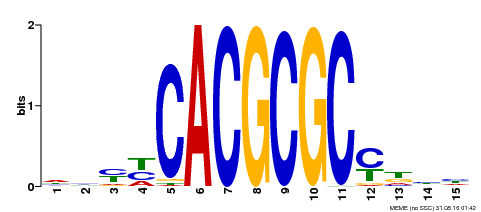
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011033967.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1-like isoform X2 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A2K2B580 | 0.0 | A0A2K2B580_POPTR; Uncharacterized protein | ||||
| TrEMBL | A0A2K2B585 | 0.0 | A0A2K2B585_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0003s10970.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF40 | 30 | 530 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||



