 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG029234.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1073aa MW: 118858 Da PI: 8.256 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.7 | 1.1e-40 | 145 | 222 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dl++ak+yhrrhkvCevhska+++lv +++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+q+
CCG029234.1 145 TCQVDNCKEDLTKAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHPLTEFDEGKRSCRRRLAGHNRRRRKTQP 222
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.9E-34 | 140 | 207 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.906 | 143 | 220 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.45E-37 | 144 | 224 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.7E-29 | 146 | 219 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1073 aa Download sequence Send to blast |
MEEVGAQVAA PIFIHQALST RYCDMTSMAK KHELSYQSPN SQLQQHQFLQ TSREKNWNSK 60 AWDWDSVGFV AKPSVAAETL RLGTVSRELK KKDKSDSKNK SNSVSEDDDG LGLNLGGSLT 120 SVEEPASRPS KRVRSGSPGN GSYPTCQVDN CKEDLTKAKD YHRRHKVCEV HSKATKALVG 180 KQMQRFCQQC SRFHPLTEFD EGKRSCRRRL AGHNRRRRKT QPEDVTSRLL LPGNRDMNNN 240 GNLDIVNLLT ALARSQGGND DKSTNCPTVP DKDQLIQILN KINSLPLPMD LAAKLSNIAS 300 LNVKNPNQPS LGHQNRLNGT ASSPSTNDLL AVLSTTLTAS APDALAILSQ RSSQSSDSDK 360 SKLPGPNQVT VPHLQKRSNF DFPAVGVERI SHCYESPAED SDYQIQESRP NLPLQLFSSS 420 PENESRQKPA SPGKYFSSDS SNPIEERSPS SSPPVVQKLF PLQSTAETMK SEKMSVSREV 480 NANVGGGRSH GSVLPLELFR GPNREPDHSS FQSFPYQGGY TSSSGSDHSP SSQNSDPQDR 540 TGRIIFKLFD KDPSHFPGTL RTKIYNWLSN SPSDMESYIR PGCVVLSVYL SMPSASWEQL 600 ERNLLQLVDS LVQDSDSDLW KSGRFLLNTG RQLASHKDGK VRLCKSWRTW SSPELILVSP 660 VAVISGQETS LQLKGRNLTG LGTKIHCTYM GGYTSKEVTD SSSPGSMYDE INVGGFKIHG 720 PSPSILGRCF IEVENGFKGN SFPVIIADAS ICKELRLLES EFDEKVLVSN IVSEEQARDF 780 GRPRSREEVM HFLNELGWLF QRKSMPSMHE VPDYSVNRFK FLLIFSVERD YCVLVKTILD 840 MLVERNTCRD ELSKEHLEML HEIQLLNRSV KRRCRKMADL LIHYYIISGD NSSRTYIFPP 900 NVGGPGGITP LHLAACASGS DGLVDALTND PHEIGLSCWN SVLDANGLSP YAYAVMTKNH 960 SHNLLVARKL AGKRNGQISV AIGNEIEQAA LEQEPMTISH FQHERKSCAK CASVAAEIHG 1020 RFLGSQGLLQ RPYIHSMLAI AAVCVCVCLF FRGAPDIGLV APFKWENLNY GTI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-31 | 136 | 219 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00664 | PBM | Transfer from Potri.002G002400 | Download |
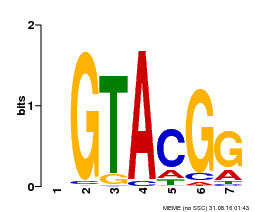
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011034771.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 isoform X1 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A2K2BB79 | 0.0 | A0A2K2BB79_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0002s00440.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9694 | 31 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



