 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | CCG029895.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 255aa MW: 29266.9 Da PI: 9.1596 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65.7 | 9.3e-21 | 96 | 146 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s y+G+r++ +g+W+AeIrdp + +r++lg++ taeeAaka+++a+k+++g
CCG029895.1 96 SVYRGIRQRT-WGKWAAEIRDPH----KgARVWLGTYNTAEEAAKAYDEAAKRIRG 146
78*****999.**********72....35************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.37E-22 | 96 | 155 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.0E-14 | 96 | 146 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.63E-30 | 96 | 154 | No hit | No description |
| SMART | SM00380 | 6.6E-37 | 97 | 160 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.2E-32 | 97 | 155 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.393 | 97 | 154 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.0E-11 | 98 | 109 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.0E-11 | 120 | 136 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008219 | Biological Process | cell death | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051707 | Biological Process | response to other organism | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MCGGAIISDF VAVKRGRRLT VEDLWSELDP FSEFLRFDHH SNDNNGSKKD PSNLLFPQKP 60 SYIIQVQQVI TEKVEKPGRA TEKGNGNKKA QRNRKSVYRG IRQRTWGKWA AEIRDPHKGA 120 RVWLGTYNTA EEAAKAYDEA AKRIRGDKAK LNFPSQTPPS TEAPPPAKKR CILAPETAYV 180 ASPFTPPLQE PYFGYQKEDY ELEEQILKLE SFLGLEHDQM AARLRENGGD YRDSGDLWML 240 DDLVTHHQYR RQINY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 3e-21 | 98 | 154 | 3 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| 2gcc_A | 3e-21 | 98 | 154 | 6 | 63 | ATERF1 |
| 3gcc_A | 3e-21 | 98 | 154 | 6 | 63 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00109 | PBM | Transfer from AT3G16770 | Download |
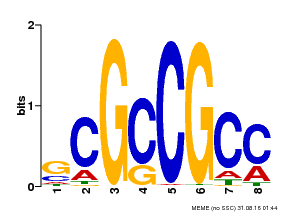
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011010669.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-3-like | ||||
| TrEMBL | U5G9D1 | 1e-170 | U5G9D1_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0008s23260.1 | 1e-171 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4892 | 33 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G16770.1 | 2e-48 | ethylene-responsive element binding protein | ||||



