 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.0053s0009.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 923aa MW: 104673 Da PI: 7.4954 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 162.5 | 7.8e-51 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseen 91
+e +rwl+++ei+a+L n++ +++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah+++n
Cagra.0053s0009.1.p 30 DEaYTRWLRPNEIHALLCNHKFFTINVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGNDN 119
45589************************************************************************************* PP
CG-1 92 ptfqrrcywlLeeelekivlvhylevk 118
ptf rrcywlL++++e+ivlvhy+e++
Cagra.0053s0009.1.p 120 PTFVRRCYWLLDKSQEHIVLVHYRETH 146
************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 76.405 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 7.1E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.3E-45 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.2E-13 | 373 | 459 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.31E-13 | 559 | 669 | No hit | No description |
| Pfam | PF12796 | 9.7E-7 | 560 | 639 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.5E-15 | 560 | 672 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.02E-15 | 568 | 679 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.292 | 577 | 672 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.7E-6 | 610 | 639 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 610 | 642 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 6.595 | 759 | 785 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 260 | 774 | 796 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.5E-4 | 797 | 819 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.237 | 798 | 822 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.2E-4 | 800 | 819 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 6.8 | 873 | 895 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.37 | 874 | 903 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.22 | 875 | 895 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 923 aa Download sequence Send to blast |
MAGVDSGKLI GSEIHGFHTL QDLDIQTMLD EAYTRWLRPN EIHALLCNHK FFTINVKPVN 60 LPKSGTIVLF DRKMLRNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGNDNP 120 TFVRRCYWLL DKSQEHIVLV HYRETHEAQA APATPGNSYS SSISDHLSPK LVAEDINSVV 180 RNTCNTGFEV RSNSLGARNH EIRLHEINTL DWDELLVPAD ISNQSHPTEE DMLYFTEQLE 240 TAPRGSAKQG NHLAGYNGSV DIPSFPGLED PVYQNNNSCG AGEFSSQHVH CGVESILQRR 300 NSNATVADQP GDALLNNGYG SQDSFGRWVN NFISDSPGSV DDPSLEAVYT PGQESSTTAV 360 SHSHSNAPEQ VFNITDVSPA WAYSTEKTKI LVTGFFHDSF QHFGRSNLFC ICGELRVPAE 420 FLQLGVYRCF LPPQSPGVVN LYLSVDGNKP VSQLFSFEHR SVPVIEKAIP QDDQLHKWEE 480 FEFQVRLAHL LFTSSNKISV LTSKISPDNL LEAKKLASRT SHLLNSWAYL MKSIQANEVP 540 FDQARDHLFE LTLKNRLKEW LLEKVIENRN TKEYDSKGLG VIHLCAVLGY TWSILLFSWA 600 NISLDFRDKH GWTALHWAAY YGREKMVAAL LSAGARPNLV TDPTKEFLGG CTPADLAQQK 660 GYDGLAAFLA EKCLVAQFKD MQVAGNISGN LETIKAEQSS NPGNANEEEQ SLKDTLAAYR 720 TAAEAAARIQ GAFREHELKV RSSAVRFASK EEEAKNIIAA MKIQHAFRNF ETRRKIAAAA 780 RIQYRFQTWK MRREFLNMRN KAIRIQAAFR GYQVRRQYQK ITWSVGVLEK AILRWRLKRK 840 GFRGLQVSQP EKKEGDEEVE DFYKTSQKQA EDRLERSVVK VQAMFRSKKA QQDYRRMKLA 900 HEEAQLEYDG MQELDQMAME ES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
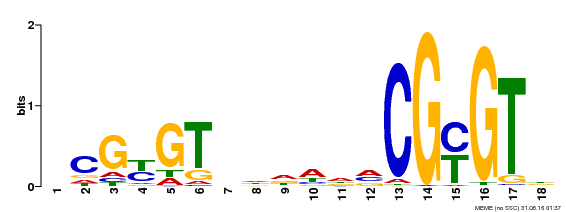
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.0053s0009.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006282550.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | R0GWD2 | 0.0 | R0GWD2_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.0053s0009.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.0053s0009.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



