 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.0053s0009.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 920aa MW: 104340 Da PI: 7.6107 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 162.5 | 7.7e-51 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseen 91
+e +rwl+++ei+a+L n++ +++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah+++n
Cagra.0053s0009.2.p 30 DEaYTRWLRPNEIHALLCNHKFFTINVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGNDN 119
45589************************************************************************************* PP
CG-1 92 ptfqrrcywlLeeelekivlvhylevk 118
ptf rrcywlL++++e+ivlvhy+e++
Cagra.0053s0009.2.p 120 PTFVRRCYWLLDKSQEHIVLVHYRETH 146
************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 76.405 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 7.1E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.3E-45 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.2E-13 | 370 | 456 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.46E-13 | 556 | 666 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.5E-15 | 557 | 669 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 9.6E-7 | 557 | 636 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.02E-15 | 565 | 676 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.292 | 574 | 669 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.7E-6 | 607 | 636 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 607 | 639 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 6.595 | 756 | 782 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 260 | 771 | 793 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.5E-4 | 794 | 816 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.237 | 795 | 819 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.2E-4 | 797 | 816 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 6.8 | 870 | 892 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.37 | 871 | 900 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.22 | 872 | 892 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 920 aa Download sequence Send to blast |
MAGVDSGKLI GSEIHGFHTL QDLDIQTMLD EAYTRWLRPN EIHALLCNHK FFTINVKPVN 60 LPKSGTIVLF DRKMLRNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGNDNP 120 TFVRRCYWLL DKSQEHIVLV HYRETHEAQA APATPGNSYS SSISDHLSPK LVAEDINSVV 180 RNTCNTVRSN SLGARNHEIR LHEINTLDWD ELLVPADISN QSHPTEEDML YFTEQLETAP 240 RGSAKQGNHL AGYNGSVDIP SFPGLEDPVY QNNNSCGAGE FSSQHVHCGV ESILQRRNSN 300 ATVADQPGDA LLNNGYGSQD SFGRWVNNFI SDSPGSVDDP SLEAVYTPGQ ESSTTAVSHS 360 HSNAPEQVFN ITDVSPAWAY STEKTKILVT GFFHDSFQHF GRSNLFCICG ELRVPAEFLQ 420 LGVYRCFLPP QSPGVVNLYL SVDGNKPVSQ LFSFEHRSVP VIEKAIPQDD QLHKWEEFEF 480 QVRLAHLLFT SSNKISVLTS KISPDNLLEA KKLASRTSHL LNSWAYLMKS IQANEVPFDQ 540 ARDHLFELTL KNRLKEWLLE KVIENRNTKE YDSKGLGVIH LCAVLGYTWS ILLFSWANIS 600 LDFRDKHGWT ALHWAAYYGR EKMVAALLSA GARPNLVTDP TKEFLGGCTP ADLAQQKGYD 660 GLAAFLAEKC LVAQFKDMQV AGNISGNLET IKAEQSSNPG NANEEEQSLK DTLAAYRTAA 720 EAAARIQGAF REHELKVRSS AVRFASKEEE AKNIIAAMKI QHAFRNFETR RKIAAAARIQ 780 YRFQTWKMRR EFLNMRNKAI RIQAAFRGYQ VRRQYQKITW SVGVLEKAIL RWRLKRKGFR 840 GLQVSQPEKK EGDEEVEDFY KTSQKQAEDR LERSVVKVQA MFRSKKAQQD YRRMKLAHEE 900 AQLEYDGMQE LDQMAMEES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
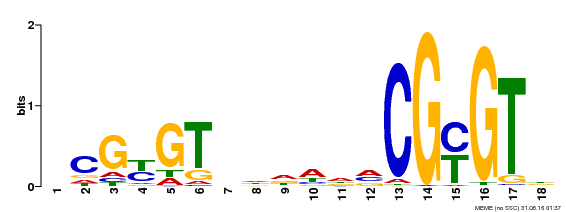
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.0053s0009.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023635492.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X3 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | R0GWD2 | 0.0 | R0GWD2_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.0053s0009.1.p | 0.0 | (Capsella grandiflora) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.0053s0009.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



