 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.0807s0010.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1374aa MW: 153923 Da PI: 7.2474 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12 | 0.00066 | 1256 | 1281 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++sF++ +L H r+ +
Cagra.0807s0010.1.p 1256 YQCDmeGCTMSFNTEKQLALHKRNiC 1281
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 15.1 | 6.7e-05 | 1281 | 1303 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H+r+H
Cagra.0807s0010.1.p 1281 CPvkGCGKNFFSHKYLVQHQRVH 1303
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 12 | 0.00066 | 1339 | 1365 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
Cagra.0807s0010.1.p 1339 YVCAepGCGQTFRFVSDFSRHKRKtgH 1365
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 6.6E-16 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.91 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 5.7E-14 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 2.3E-53 | 200 | 369 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 34.846 | 203 | 369 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 3.57E-28 | 216 | 387 | No hit | No description |
| Pfam | PF02373 | 2.0E-38 | 233 | 352 | IPR003347 | JmjC domain |
| SMART | SM00355 | 9.1 | 1256 | 1278 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.01 | 1279 | 1303 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.173 | 1279 | 1308 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-6 | 1280 | 1307 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1281 | 1303 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.72E-10 | 1295 | 1337 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.7E-9 | 1308 | 1332 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.928 | 1309 | 1338 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0015 | 1309 | 1333 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1311 | 1333 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.14E-8 | 1327 | 1361 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.7E-9 | 1333 | 1362 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.032 | 1339 | 1370 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.62 | 1339 | 1365 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1341 | 1365 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1374 aa Download sequence Send to blast |
MAVSEQSQDV FPWLKSLPVA PEFRPTLAEF QDPIAYIFKI EEEASKYGIC KILPPVPPPS 60 KKTSISNLNR SLAARAAARV RDGGFGACDY DGGPTFATRQ QQIGFCPRKQ RPVQRPVWQS 120 GEEYSFGEFE FKAKSFEKAY LKKCGKKSQL SALEIETLYW RATVDKPFSV EYANDMPGSA 180 FIPLSLAAAR RRESGGDGGT VGETAWNMRA MARAEGSLLK FMKEDIPGVT SPMVYIAMMF 240 SWFAWHVEDH DLHSLNYLHM GASKTWYGVP KEAAPAFEEV VRVHGYGGEL NPLVTFSTLG 300 EKTTVMSPEV FVKAGIPCCR LVQNPGEFVV TFPGAYHSGF SHGFNFGEAS NIATPQWLRM 360 AKDAAIRRAA INYPPMVSHL QLLYDFALAL GSRVPTSINA KPRSSRLKDK KRSEGERLTK 420 KLFVDNIIRN NELLYSLGKG SPVALLPQSS SDISVCSDLR IGSDLRTNQE SPIQLKSEDL 480 SCDSVMVGLN NGLKDTVSVK EKFTSLCERN RNHLASRGKE TQETLSDTEG RNNAGAVGLS 540 DQRLFSCVTC GVLSFDCVGI VQPKEASAGY IMSANSSFFN DWTAASGSAN RGQDARSLHL 600 QSTEKDDVNY FSNVPVQTGH QRTLTTSLTN AHKDNGALGL LASAYGDSSD SEEDDNKGFD 660 ASDLEGERKK YDQESACAFK ASSFDTDGNE EARDGQTSDF NSQRLTSKQN SLSKCGISSL 720 LEITLPFIPR SDDDPCRLHV FCLEHAAEVE QQLRSIGGIH IMLLCHPEYP RIEAEAKIVA 780 EEFAINHEWN DTEFRNVTRE DEETIQAALD NVEAKGGNSD WTVKLGINLS YSAFLSRSPL 840 YSKQMPYNSV IYNVFGRSST AVSSLSKPEV SGRRSSRQRK YVVGKWCGKV WMSHQVHPFL 900 LEEDLEGEES ERSCPLRAAM DEDVTGKRLF SCNVSRDAAT MFGRKYCRKR KIRAKAVPRK 960 KLTAFKREDG VSDDTSEDHS YKQQWRASGN EEESYFETGH TASGDSSNQM SDQGKEIVRH 1020 KGYNGFESDD EVSDRSLGEE YTVRECAVSE SSMENGFQPY REKQAMYDDN DDDGDDDDDD 1080 DMYRHPRGIP RSKRSKVIRN PVSYDSEEND VYQQRGRRVS RSSRQANRMG GEYDSAENSF 1140 EELDFCTTGK RQTRSTAKRK AKTETVQSPR DTKGRSLQEF ASGKKIEELD SYMEGPSTRL 1200 RLRNQKPSRG SFESKPKKVG KKRSSNASFS RAASEEYVQE DEEAEAENEE EECTGYQCDM 1260 EGCTMSFNTE KQLALHKRNI CPVKGCGKNF FSHKYLVQHQ RVHSDDRPLK CPWKGCKMTF 1320 KWAWSRTEHI RVHTGARPYV CAEPGCGQTF RFVSDFSRHK RKTGHSVKKT KKR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 3e-77 | 1253 | 1373 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 3e-77 | 1253 | 1373 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 3e-77 | 1253 | 1373 | 20 | 140 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 948 | 960 | KRKIRAKAVPRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
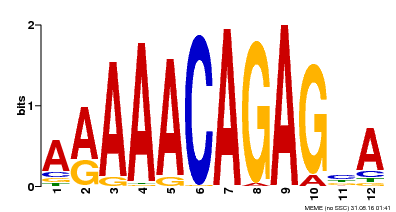
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.0807s0010.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY664499 | 0.0 | AY664499.1 Arabidopsis thaliana relative of early flowering 6 (REF6) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023637886.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | R0FN14 | 0.0 | R0FN14_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.0807s0010.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.0807s0010.1.p |



