 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.1305s0026.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 455aa MW: 50080.7 Da PI: 9.0664 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 44.3 | 3.9e-14 | 374 | 423 | 5 | 54 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkk 54
+r++r++kNRe+A rsR+RK+a++ eLe ++++L++ N++L+k+ e+ +
Cagra.1305s0026.1.p 374 RRQKRMIKNRESAARSRARKQAYTMELEAEIAQLKELNEELQKKQVEIME 423
69*************************************99876555444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 9.4E-14 | 370 | 435 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.473 | 372 | 417 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.88E-10 | 374 | 426 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.3E-13 | 374 | 430 | No hit | No description |
| Pfam | PF00170 | 7.6E-12 | 374 | 430 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 1.33E-26 | 374 | 428 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 377 | 392 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 455 aa Download sequence Send to blast |
MGSRTDFKNF VDGLTDEAAI HPKQPMMGTT LPLTRQNSVF SLTFDEFQNS WGGGIGKDFG 60 SMNMDELLKN IWTAEESHSM MGNNTSFNNI NNGNALINNN GGLSVGVGGE TGGGFLTGGS 120 LQRQGSLTLP RIISQKTVDD VWKELMKEDD VGNVVGNGGT SGIPQRQQTL GEMTLEEFLL 180 RAGVVREEPQ PVESVNNFNA GFYGFGSNAG LGTARNGIVA NQPHDFAGNG AVVRPDLLTT 240 QTQPLQMQQP LQMQQPQKVH QPQQLIQKQE RPFPKQTTIA FSNTVDAVNR SQLATQFQEV 300 KPSILGMQDH HMNNNLLQAV DFKTGVTVAA VSPGSQMSPD LTPKSALDAS LSPAPYMFGR 360 VRKTGAVLEK VIERRQKRMI KNRESAARSR ARKQAYTMEL EAEIAQLKEL NEELQKKQVE 420 IMEKQKNQLL EPLRQPWGMG CKRQCLRRTL TGPW* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. {ECO:0000269|PubMed:11884679, ECO:0000269|PubMed:15361142}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
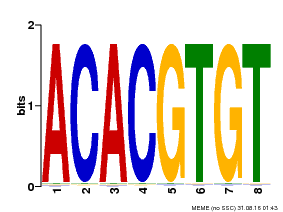
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.1305s0026.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA). {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:16284313}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF093546 | 0.0 | AF093546.1 Arabidopsis thaliana clone 11 abscisic acid responsive elements-binding factor (ABRE) mRNA, complete cds. | |||
| GenBank | AK175851 | 0.0 | AK175851.1 Arabidopsis thaliana mRNA for abscisic acid responsive elements-binding factor (ABRE/ABF3), complete cds, clone: RAFL22-47-B14. | |||
| GenBank | AY054605 | 0.0 | AY054605.1 Arabidopsis thaliana Unknown protein (F17I5.190:F17I5.200) mRNA, complete cds. | |||
| GenBank | AY081467 | 0.0 | AY081467.1 Arabidopsis thaliana unknown protein (F17I5.190:F17I5.200) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006284949.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 6 isoform X1 | ||||
| Refseq | XP_023635587.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 6 isoform X1 | ||||
| Refseq | XP_023635588.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 6 isoform X1 | ||||
| Swissprot | Q9M7Q3 | 0.0 | AI5L6_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ||||
| TrEMBL | R0F7H3 | 0.0 | R0F7H3_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.1305s0026.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM734 | 27 | 129 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34000.2 | 0.0 | abscisic acid responsive elements-binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.1305s0026.1.p |



