 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.1317s0005.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 494aa MW: 54195 Da PI: 7.5008 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.2 | 7.6e-17 | 166 | 215 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Cagra.1317s0005.1.p 166 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAVKFRG 215
78*******************......55***********************9997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 2.53E-11 | 166 | 222 | No hit | No description |
| SuperFamily | SSF54171 | 2.42E-15 | 166 | 223 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.2E-8 | 166 | 215 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.359 | 167 | 223 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.1E-16 | 167 | 223 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.3E-30 | 167 | 229 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 494 aa Download sequence Send to blast |
MLDLNLDVDS TESTQNGRDS VTVKGVSLNQ MDESVTSNSS VVNAEASSCI DGEEELCSTR 60 KSRTVKFQFE ILKGGGEEDE GEDDDNDDRS ALMMTKDFFP VPRDGINFMD SSAQSSRSTV 120 DISFQRGKQG GDFVGSGSGG GDAARVMQPP SQPVKKSRRG PRSKSSQYRG VTFYRRTGRW 180 ESHIWDCGKQ VYLGGFDTAH AAARAYDRAA VKFRGLEADI NFIISDYEED LKQMANLSKE 240 EVVQVLRRQS SGFSRNNSRY QGVALQKIGG WGAHMEQFHG NMACDKAAIQ WNGRQAASFI 300 EPHGSRMIPE AANVKLDLNL GISLSLGDGP KQKDRTHRLH HVPNSIICGR NTKMENHMAA 360 ASCDTPFNFL KRGSDHLNNR QALPSAFFSP MERTPEKGPM LRSHQSFPAR TWQGHDQSSS 420 GTAVAATAPQ LFSNAASSGF SLSDTRPPSS PATPQSSQPF VNLNHPGLYV IHPSDYTTQH 480 HHHNLMNRSQ PPP* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00616 | PBM | Transfer from AT5G60120 | Download |
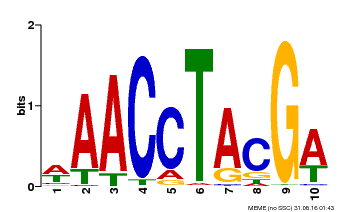
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.1317s0005.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT001916 | 0.0 | BT001916.1 Arabidopsis thaliana clone C105157 putative APETALA2 protein (At5g60120) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006280364.1 | 0.0 | AP2-like ethylene-responsive transcription factor TOE2 | ||||
| Swissprot | Q9LVG2 | 0.0 | TOE2_ARATH; AP2-like ethylene-responsive transcription factor TOE2 | ||||
| TrEMBL | R0EVS3 | 0.0 | R0EVS3_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.1317s0005.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM12836 | 17 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G60120.1 | 0.0 | target of early activation tagged (EAT) 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.1317s0005.1.p |



