 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.16630s0008.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 935aa MW: 103951 Da PI: 5.7037 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 133.9 | 5.1e-42 | 144 | 221 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv +Ceadls++k+yhrrhkvCe+hska++++v g++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Cagra.16630s0008.1.p 144 VCQVGNCEADLSKVKDYHRRHKVCEMHSKATSAIVGGIMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 221
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.7E-33 | 140 | 206 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.274 | 142 | 219 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.36E-39 | 143 | 223 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.8E-30 | 145 | 218 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 9.2E-8 | 724 | 826 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 5.6E-8 | 724 | 825 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.83E-9 | 727 | 823 | No hit | No description |
| PROSITE profile | PS50297 | 8.561 | 727 | 823 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 935 aa Download sequence Send to blast |
MEARIEGEPQ HFYGFRAVDL RSVGKRTVEW DLNDWKWDGD LFVATQLHPG GSETTGRQFF 60 PIGNSSNSSS SCSDEAHNSE SLVRSTREVE KKRRAVAIEG DTNGGSLTLK LGDNGCDLNG 120 EREGNSAAKK TKFGGAGAVT NRAVCQVGNC EADLSKVKDY HRRHKVCEMH SKATSAIVGG 180 IMQRFCQQCS RFHVLQEFDE GKRSCRRRLA GHNKRRRKTN PEPGANGNPS DDQSSNYLLI 240 SLLKILSNMH TTRSDHTGDQ DLMSHLLKSL VSHAGEQLGK NLVELLLQGG SQGSLNLGNS 300 GLLAIEEAPQ EDLKQLSVNV PEIPQQELYA NGTAAENRSE KQVRMNDFDL NDIYIDSDDT 360 DIERSPPPTN PATSSLDYPS WIHQSSPPQT SRNSDSASDQ SPSSSGEDAQ MRTGRIVFKL 420 FGKEPNDFPV VLRGQILDWL SHSPTDMESY IRPGCIVLTI YLRQAETAWE ELSDDLGFSL 480 RKLLDLSDDP LWTTGWIYVR VQNQLAFVYN GQVVVDTSLP LKSHDYSHII SVKPLAVAAA 540 GKAQFTVKGI NLRRRGTRLL CAVEGKYLIQ ESTHDSTTEE NDDQNNEIVE CVNFSCDLPI 600 ISGRGFMEIE DQGLSSSFFP FLVVEDNDVC SEIRILETTL EFTGTDSAKQ AMDFIHEIGW 660 LLHRSKLGES DPNPDVFPLI RFQWLIEFSM DREWCAVIRK LLNMFFDGAV GDFSSSSNAT 720 LSELCLLHRA VRKNSKPLVE MLLNYVPKQQ RNSLFRPDAA GPAGLTPLHI AAGKDGSEDV 780 LDALTEDPAM VGIEAWKTSR DSTGFTPEDY ARLRGHFSYI HLIQRKINKK STAEDHVVVN 840 IPVSFSDREQ KEPKSGPVTS ALEITQINQL PCKLCDHKLV YGTTRRSVAY RPAMLSMVAI 900 AAVCVCVALL FKSCPEVLYV FQPFRWELLD YGTS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 9e-32 | 135 | 218 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
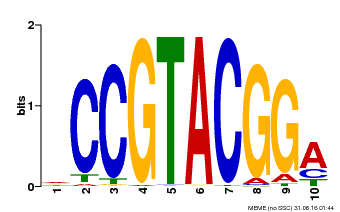
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.16630s0008.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC815948 | 0.0 | KC815948.1 Arabis alpina SQUAMOSA promoter binding protein-like protein 1 (SPL1) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023639628.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Refseq | XP_023639629.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | D7LFY4 | 0.0 | D7LFY4_ARALL; Uncharacterized protein | ||||
| STRING | Cagra.16630s0008.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.16630s0008.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



