 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.23502s0003.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 331aa MW: 37858 Da PI: 6.977 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 96.2 | 2.1e-30 | 37 | 122 | 1 | 84 |
EEEE-..-HHHHTT-EE--HHH.HTT...---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh...ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkl 84
f+kvltpsdv+k++rlv+pk++ae++ + + ++++ l ++d++gr W+++++y+++s++yv+tkGW++Fvk+++L +gD+v F++
Cagra.23502s0003.1.p 37 FDKVLTPSDVGKLNRLVIPKQHAENYfplEDN-QNGTVLDFQDRNGRMWRFRYSYWNSSQSYVMTKGWSRFVKDKKLTSGDTVSFHR 122
89*********************999**7444.46789***********************************************93 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 3.9E-38 | 32 | 122 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.33E-26 | 35 | 122 | No hit | No description |
| SuperFamily | SSF101936 | 1.28E-29 | 35 | 122 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 5.1E-27 | 37 | 122 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 6.5E-22 | 37 | 141 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.536 | 37 | 141 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MNMSTMNSEQ ELTENGAGGS GGGNNYFYTA EMREHMFDKV LTPSDVGKLN RLVIPKQHAE 60 NYFPLEDNQN GTVLDFQDRN GRMWRFRYSY WNSSQSYVMT KGWSRFVKDK KLTSGDTVSF 120 HRGFITDDNA PERRLKIMFI DWRHRDTHNN YGFGSPPYPT ASFYPPPQYP MPLPSYRSFP 180 PFYHNQYQER EYLGYGYGRL VNGGRYYAGT PLDHHHHHQW SLGRSQPLVY ESVPVFPAAR 240 APPPPPAPPQ PPITRRLRLF GVDVEESSPY VETRGEMGVA GHSSSSPVVI GDDDQLSWRL 300 PSGEVDASTS LPMQLSDDED YKRKGKFSEY * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5os9_A | 2e-48 | 33 | 145 | 4 | 113 | B3 domain-containing transcription factor NGA1 |
| 5os9_B | 2e-48 | 33 | 145 | 4 | 113 | B3 domain-containing transcription factor NGA1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Regulates lateral organ growth. Functionally redundant with NGA1, NGA2 and NGA3. {ECO:0000269|PubMed:16603651}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00424 | DAP | Transfer from AT4G01500 | Download |
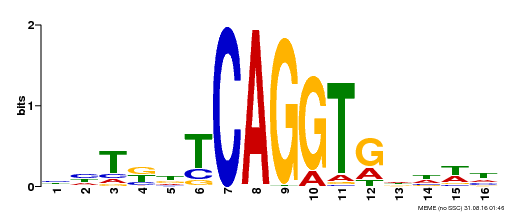
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.23502s0003.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006288169.1 | 0.0 | B3 domain-containing transcription factor NGA4 | ||||
| Swissprot | O82595 | 1e-170 | NGA4_ARATH; B3 domain-containing transcription factor NGA4 | ||||
| TrEMBL | R0FFQ7 | 0.0 | R0FFQ7_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.23502s0003.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13651 | 15 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01500.1 | 1e-148 | B3 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.23502s0003.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



