 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.3356s0080.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 612aa MW: 66705.5 Da PI: 4.5281 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.4 | 1.1e-12 | 438 | 483 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L
Cagra.3356s0080.1.p 438 NHVEAERQRREKLNQRFYSLRAVVPNV-----SKMDKASLLGDAISYINEL 483
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 3.2E-55 | 63 | 259 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.282 | 434 | 483 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.31E-18 | 437 | 504 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.70E-15 | 437 | 488 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 6.6E-19 | 438 | 504 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.0E-10 | 438 | 483 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 9.0E-17 | 440 | 489 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 612 aa Download sequence Send to blast |
MSPTSVQITD YHLNQSTNGT TNLWSNDDDA SVMEAFIGGS DQSSLFPPSP PPPAQSQFNE 60 DTLQQRLQAL IEGANESWTY AVFWQSSYDF AGEDDGGGES RNTAVLGWGD GYYKGEEENS 120 RKKKSNPASA AEQEHRRRVI RELNALISGG GGVVNNGGGS DEAGDEEVTD TEWFFLVSMT 180 QSFVSGTGLP GQAFSNSNTI WLSGSNALAG SSCERARQGQ IYGLQTMVCV PCENGVVELG 240 SSEIIHQSSD LVDKVDTFFN FNNGGGESGS WAFNLNPDQG ENDPGLWIGE PDSVGVELGL 300 VAPVMNNTGN NSASNSDSQP ISKLCNGSSV EDPKPQVTKS SEMVSFKNGT DENGFSGQSR 360 FMEEDSNKKR SPVSNNDEGM LSFTSVLPRP AKSGDSNHSD LEASVVKEAE SNRTVVEPEK 420 KPRKRGRKPA NGREEPLNHV EAERQRREKL NQRFYSLRAV VPNVSKMDKA SLLGDAISYI 480 NELKSKLQKV ESDKEELQKQ IEGMSKEAAN EKSSVKERKC ANQESGVSIE MEVDVKIIGW 540 DAMIRVQCSK RNHPGAKFME ALKELDLEVN HASLSVVNDL MIQQATVKMG REFFTQDQLK 600 VALMEKVGEC L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rru_A | 6e-93 | 20 | 255 | 3 | 227 | Transcription factor MYC3 |
| 4ywc_A | 6e-93 | 20 | 255 | 3 | 227 | Transcription factor MYC3 |
| 4ywc_B | 6e-93 | 20 | 255 | 3 | 227 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 419 | 427 | KKPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in jasmonic acid (JA) gene regulation. With MYC2 and MYC3, controls additively subsets of JA-dependent responses. Can form complexes with all known glucosinolate-related MYBs to regulate glucosinolate biosynthesis. Binds to the G-box (5'-CACGTG-3') of promoters. Activates multiple TIFY/JAZ promoters. {ECO:0000269|PubMed:21321051, ECO:0000269|PubMed:21335373, ECO:0000269|PubMed:23943862}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00086 | PBM | Transfer from AT4G17880 | Download |
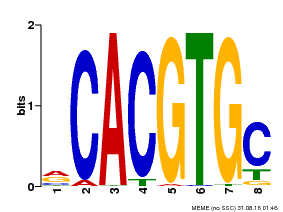
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.3356s0080.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. Not induced by jasmonic acid. {ECO:0000269|PubMed:12679534, ECO:0000269|PubMed:21335373}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF251689 | 0.0 | AF251689.1 Arabidopsis thaliana putative transcription factor BHLH4 (BHLH4) mRNA, complete cds. | |||
| GenBank | AK221507 | 0.0 | AK221507.1 Arabidopsis thaliana mRNA for putative transcription factor BHLH4, complete cds, clone: RAFL07-57-K04. | |||
| GenBank | AL021889 | 0.0 | AL021889.2 Arabidopsis thaliana DNA chromosome 4, BAC clone T6K21 (ESSA project). | |||
| GenBank | AL161547 | 0.0 | AL161547.2 Arabidopsis thaliana DNA chromosome 4, contig fragment No. 47. | |||
| GenBank | CP002687 | 0.0 | CP002687.1 Arabidopsis thaliana chromosome 4 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006283348.1 | 0.0 | transcription factor MYC4 | ||||
| Swissprot | O49687 | 0.0 | MYC4_ARATH; Transcription factor MYC4 | ||||
| TrEMBL | R0F4A8 | 0.0 | R0F4A8_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.3356s0080.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1265 | 28 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17880.1 | 0.0 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.3356s0080.1.p |



