 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.3745s0024.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 691aa MW: 74714.4 Da PI: 6.5137 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 100.3 | 1.2e-31 | 271 | 328 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK vkgse+prsYY+Ct ++C+vkkkvers+ +++++ei+Y+g Hnh+k
Cagra.3745s0024.1.p 271 EDGYNWRKYGQKLVKGSEYPRSYYKCTNPNCQVKKKVERSR-EGHITEIIYKGAHNHSK 328
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 103.8 | 9.2e-33 | 491 | 549 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct +gC+v+k+ver+++d k v++tYeg+Hnh+
Cagra.3745s0024.1.p 491 LDDGYRWRKYGQKVVKGNPNPRSYYKCTAPGCTVRKHVERASHDLKSVITTYEGKHNHD 549
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.8E-28 | 263 | 329 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.97E-25 | 268 | 329 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.6E-34 | 270 | 328 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.9E-24 | 271 | 326 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.25 | 271 | 329 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.0E-36 | 476 | 551 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.57E-28 | 483 | 551 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.136 | 486 | 551 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.8E-37 | 491 | 550 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.9E-25 | 492 | 549 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 691 aa Download sequence Send to blast |
MAGFDKNIAV MGEWVPRSRS PSPGELLSSA IGEGKGTKRV LERELSLNHG QVTGVEVDTT 60 GNHNKDSSQT NVSRGCLSER IAARAGFNAP RLNTENIRSN TNYSIDSSPC LTISSPGLSP 120 ATLLESPVFL SNPMAQPSPT TGKFPFLPGF NSNALSSDKA KDEFFDDIGA SFTFQSVSRS 180 SSSFFQGTTE MMSAEYGNYN NRSSHQPADE DVKPGSQNIE SSNLYGIEFD PSGQNKTSDA 240 TTNTGLETVD HQEEEEDQRR GDSMVGGAPA EDGYNWRKYG QKLVKGSEYP RSYYKCTNPN 300 CQVKKKVERS REGHITEIIY KGAHNHSKPL PNRRSGMQTD GTEQVEQQQQ QQQQQRDSAG 360 TWVSCNNAQQ QGGSNVNNVE ERSTRFEYGN QSGVIQAQTG GQYDSGDAVV VVDTSSTFSN 420 DEDEDDRGTH GSVSLGCDGG AGGGGGGGGE GEESESKRRK LEAYAAEMSG ATRAIREPRV 480 VVQTTSDVDI LDDGYRWRKY GQKVVKGNPN PRSYYKCTAP GCTVRKHVER ASHDLKSVIT 540 TYEGKHNHDV PAARNSSHGG GGGSSGGSAA ASHHYNNVHQ SEQSRGRFDR QVTTTHHQSP 600 YGRPFTFQPH LGPPSGYAFG LGQNGLANLS MPGLAFGQGK MPGLPHQYLT QQPVGMNEAM 660 MQRGMEPKAE PVSDSGQSVY NQIMSRLPQI * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 5e-37 | 271 | 552 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 5e-37 | 271 | 552 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
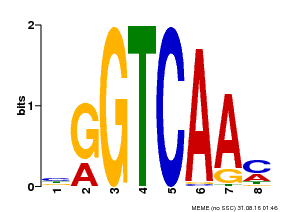
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.3745s0024.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF418308 | 0.0 | AF418308.1 Arabidopsis thaliana WRKY transcription factor 2 (WRKY2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006280109.1 | 0.0 | probable WRKY transcription factor 2 | ||||
| Swissprot | Q9FG77 | 0.0 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | R0EV69 | 0.0 | R0EV69_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.3745s0024.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7954 | 27 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56270.1 | 0.0 | WRKY DNA-binding protein 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.3745s0024.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



