 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.4093s0003.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 987aa MW: 109649 Da PI: 5.9075 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.9 | 4.8e-18 | 31 | 77 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde l +av+++ g++Wk+Ia+++ Rt+ qc +rwqk+l
Cagra.4093s0003.1.p 31 KGQWTPEEDEVLCKAVERFQGKNWKKIAECFK-DRTDVQCLHRWQKVL 77
799****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 58.5 | 1.5e-18 | 83 | 129 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEd ++ +v+++G + W+tI+++++ gR +kqc++rw+++l
Cagra.4093s0003.1.p 83 KGPWSKEEDNTIIALVERYGAKKWSTISQHLP-GRIGKQCRERWHNHL 129
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 53.6 | 5e-17 | 135 | 177 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+++WT+eE++ l++a++ +G++ W+ ++ ++ gR+++++k++w+
Cagra.4093s0003.1.p 135 KNAWTQEEELTLIRAHQIYGNK-WAELMKLLP-GRSDNSIKNHWN 177
689*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.523 | 26 | 77 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.61E-18 | 30 | 91 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-14 | 30 | 79 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.4E-15 | 31 | 77 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-23 | 33 | 94 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.81E-13 | 34 | 77 | No hit | No description |
| PROSITE profile | PS51294 | 31.363 | 78 | 133 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.74E-30 | 80 | 176 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-17 | 82 | 131 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.4E-17 | 83 | 129 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.08E-15 | 85 | 129 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 9.7E-26 | 95 | 137 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.453 | 134 | 184 | IPR017930 | Myb domain |
| SMART | SM00717 | 7.0E-15 | 134 | 182 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.9E-15 | 135 | 178 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.11E-11 | 137 | 177 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-20 | 138 | 184 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 987 aa Download sequence Send to blast |
MKAPIIPPEG FQSELKGKHG RTSGPARRST KGQWTPEEDE VLCKAVERFQ GKNWKKIAEC 60 FKDRTDVQCL HRWQKVLNPE LVKGPWSKEE DNTIIALVER YGAKKWSTIS QHLPGRIGKQ 120 CRERWHNHLN PLINKNAWTQ EEELTLIRAH QIYGNKWAEL MKLLPGRSDN SIKNHWNSSV 180 KKKLDSYYAS GLLDQCQSSP LIALQNKSIA SSSSWMYSSG DEGNSRAGVD AEESECSQAS 240 TVFSQSTNDL QEEVKPRNEE FYMPEFHSRA EHQISNDAEP YYPSFEEVKI VVPEISCEAE 300 CSKKFQNLTC SHEVRTTTAT EDQLQGMSNE AKQDPDLDLL TDNMDNDGKN QAHRQAFQSS 360 GRLSEQPSLP NSDTDPEAQT LITDEECCRV LFPDTMKDSC TSSIKQGRNM IDPRKGKGCL 420 CSQSVESHAH ETGKLPALSR HPSSSEVQAG HNCVPLSDSD LKDPVLLRND SNPPIQGSHL 480 FGATELECKT VTNDSFIDTD GHVTSHGNDN DGIEQHGLSY IPKDSLKLVP LNSFSSPSRV 540 NRIYFPIDDK PAEKDKGALC YEPPRFPSAD IPFFSCDLVP SNSDLRQEYS PFGIRQLMIS 600 SMNCTTPLRL WDSPCHERSP DVMLNDATKS LSNAPSILKK RHRDLLSPVL DRRKDKKLKR 660 AATSSLAKDL SCLDVMLDGG DDCMSTRPSE SPENKNRHAS PSKAKDNRYS ASGWSDQEMI 720 PIDEEPKETL KLGGVTSMQN QKGCNEGGAS AKNDQETSGT FVEIQLCSPG MTGGRSDNKV 780 NTSAKDLSNQ HKRSLADIPT EEISPELPCT VDSSPLSAID KTNTTETSFD IENFNIFDGT 840 PYRKLLDTPS PWKSPLLFGS FLQSPKLPPE VTFEDIGCFM SPGERSYDAI GLMKHLSEHS 900 ATAYADALEV LGNDTPETIL KKRQLNKSIQ GKENQHHPHD QLGNRSQVER RTLDFSDCGT 960 PGKSTVSSAP PGGYSSPSSY LLKSCR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 3e-65 | 31 | 184 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 3e-65 | 31 | 184 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 650 | 657 | DRRKDKKL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (PubMed:21862669). Transcription activator involved in the regulation of cytokinesis, probably via the activation of several G2/M phase-specific genes transcription (e.g. KNOLLE) (PubMed:17287251, PubMed:21862669). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). Required for the maintenance of diploidy (PubMed:21862669). {ECO:0000269|PubMed:17287251, ECO:0000269|PubMed:21862669, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00466 | DAP | Transfer from AT4G32730 | Download |
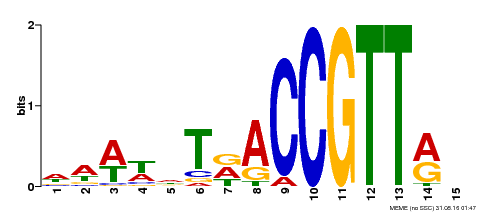
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.4093s0003.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Constant levels during cell cycle. Activated by CYCB1. {ECO:0000269|PubMed:17287251}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF151646 | 0.0 | AF151646.1 Arabidopsis thaliana PC-MYB1 (PC-MYB1) mRNA, complete cds. | |||
| GenBank | AF176005 | 0.0 | AF176005.2 Arabidopsis thaliana putative c-myb-like transcription factor (AtMYB3R-1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006283062.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Refseq | XP_023634615.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Refseq | XP_023634616.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Refseq | XP_023634617.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Swissprot | Q9S7G7 | 0.0 | MB3R1_ARATH; Transcription factor MYB3R-1 | ||||
| TrEMBL | R0GUI6 | 0.0 | R0GUI6_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.4093s0003.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4674 | 26 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32730.2 | 0.0 | Homeodomain-like protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.4093s0003.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



