 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.4138s0007.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 232aa MW: 25293.5 Da PI: 9.9893 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.3 | 6.5e-12 | 92 | 136 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ ++ + ++lG+g+W+ I+r + ++++ q+ s+ qk+
Cagra.4138s0007.1.p 92 PWTEEEHRTFLIGLEKLGKGDWRGISRNFVVTKSPTQVASHAQKH 136
8******************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.532 | 3 | 18 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 17.873 | 84 | 141 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.87E-15 | 87 | 141 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-8 | 89 | 139 | IPR001005 | SANT/Myb domain |
| TIGRFAMs | TIGR01557 | 2.4E-16 | 89 | 140 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-10 | 89 | 136 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 8.7E-10 | 92 | 136 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.98E-8 | 92 | 137 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 232 aa Download sequence Send to blast |
MGRRCSHCGN VGHNSRTCSS YHTRAIRLFG VHLDTTSSSP PPPSPPSILT AAMKKSFSMD 60 CLPACSSSSS SFAGYLSDGL SHKTPDRRKG VPWTEEEHRT FLIGLEKLGK GDWRGISRNF 120 VVTKSPTQVA SHAQKHFLRQ TTTLNHKRRR TSLFDMVSAG KVEENSTLQS ICNDHIESTS 180 KVVWKQGLLN PSLGYQDPKV SESGNSGGLD LELKLASIQS PESNIRPISV T* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds to 5'-TATCCA-3' elements in gene promoters. Contributes to the sugar-repressed transcription of promoters containing SRS or 5'-TATCCA-3' elements. Transcription repressor involved in a cold stress response pathway that confers cold tolerance. Suppresses the DREB1-dependent signaling pathway under prolonged cold stress. DREB1 responds quickly and transiently while MYBS3 responds slowly to cold stress. They may act sequentially and complementarily for adaptation to short- and long-term cold stress (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
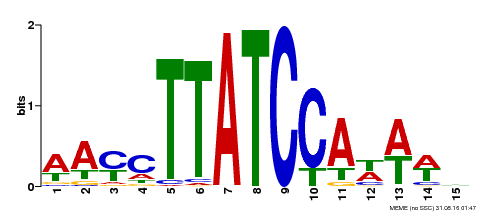
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.4138s0007.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by sucrose and gibberellic acid (GA) (PubMed:12172034). Induced by cold stress in roots and shoots. Induced by salt stress in shoots. Down-regulated by abscisic aci (ABA) in shoots (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY519517 | 0.0 | AY519517.1 Arabidopsis thaliana MYB transcription factor (At5g56840) mRNA, complete cds. | |||
| GenBank | BT012528 | 0.0 | BT012528.1 Arabidopsis thaliana At5g56840 gene, complete cds. | |||
| GenBank | BT014956 | 0.0 | BT014956.1 Arabidopsis thaliana At5g56840 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006280948.2 | 1e-169 | transcription factor MYBS3 | ||||
| Swissprot | Q7XC57 | 1e-46 | MYBS3_ORYSJ; Transcription factor MYBS3 | ||||
| TrEMBL | R0EY53 | 1e-167 | R0EY53_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.4138s0007.1.p | 1e-171 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5496 | 27 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 1e-137 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.4138s0007.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



