 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cagra.4538s0003.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 320aa MW: 34363.8 Da PI: 10.095 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.4 | 7.7e-39 | 77 | 135 | 4 | 62 |
zf-Dof 4 kalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
lkcprCds+ntkfCy+nny+l+qPr+fCkaCrryWt+GGalrnvPvGgg+r+nkk +
Cagra.4538s0003.2.p 77 GVLKCPRCDSANTKFCYFNNYNLTQPRHFCKACRRYWTRGGALRNVPVGGGCRRNKKGK 135
5789***************************************************9865 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 7.0E-37 | 60 | 130 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.2E-32 | 78 | 133 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.683 | 79 | 133 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 81 | 117 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 320 aa Download sequence Send to blast |
MSSTNSNNHP HHLQLQESGS LVSAHHQVLS HHFPQNPNPN HHHVDTTVAA ATIDPGSLNG 60 QAAERARLAK NHQPPEGVLK CPRCDSANTK FCYFNNYNLT QPRHFCKACR RYWTRGGALR 120 NVPVGGGCRR NKKGKSVSSK SSSSSSQNKP STSMVNATSP TTTSNVHLQP NSQFPFLPTL 180 QNLTHLGGIG LNLAPMNGNT SSSSFLNDLG FYHGGNTSGP VMSNNNNNEN NNIMTSLGTA 240 NNNFALFDRT MGLFNFPNES NMGLSSSGAT RASQTAPVKM EDISRSVSGL TSQGNQSNQY 300 WTGTGLPGSS SNNHHQHLM* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00281 | DAP | Transfer from AT2G28810 | Download |
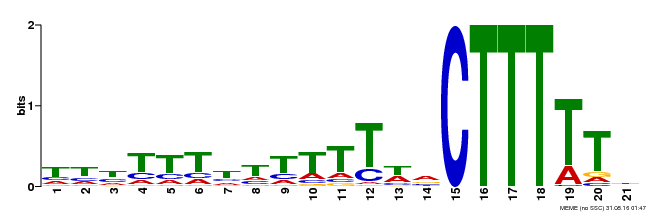
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cagra.4538s0003.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC005727 | 0.0 | AC005727.3 Arabidopsis thaliana chromosome 2 clone F8N16 map mi54, complete sequence. | |||
| GenBank | BT032877 | 0.0 | BT032877.1 Arabidopsis thaliana unknown protein (At2g28810) mRNA, complete cds. | |||
| GenBank | CP002685 | 0.0 | CP002685.1 Arabidopsis thaliana chromosome 2, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023640027.1 | 0.0 | dof zinc finger protein DOF2.2 isoform X2 | ||||
| Swissprot | Q9ZV33 | 1e-170 | DOF22_ARATH; Dof zinc finger protein DOF2.2 | ||||
| TrEMBL | R0HD44 | 0.0 | R0HD44_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.4538s0003.1.p | 0.0 | (Capsella grandiflora) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28810.2 | 1e-151 | Dof family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cagra.4538s0003.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



