 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10000120m | ||||||||
| Common Name | CARUB_v10000120mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1029aa MW: 116114 Da PI: 6.1248 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 186.2 | 3.3e-58 | 21 | 138 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
l+e ++rwl++ ei++iL+n++k+++++e++trp sgsl+L++rk++ryfrkDG++w+kkkdgkt++E+hekLKvg+++vl+cyYah+e n++f
Carubv10000120m 21 LSEaQHRWLRPAEICEILQNHHKFHIASESPTRPASGSLFLFDRKVLRYFRKDGHNWRKKKDGKTIKEAHEKLKVGSIDVLHCYYAHGEGNENF 114
45559***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrrcyw+Le++l +iv+vhylevk
Carubv10000120m 115 QRRCYWMLEQDLMHIVFVHYLEVK 138
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 85.154 | 17 | 143 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.4E-85 | 20 | 138 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 7.7E-51 | 23 | 137 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 5.29E-14 | 434 | 520 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 1.4E-7 | 616 | 694 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.5E-18 | 617 | 729 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.837 | 617 | 728 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.64E-19 | 618 | 728 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.96E-14 | 623 | 726 | No hit | No description |
| SMART | SM00248 | 3500 | 634 | 663 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.656 | 634 | 666 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 667 | 696 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.22 | 667 | 699 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.51 | 842 | 864 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 6.37E-8 | 842 | 892 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.73 | 843 | 872 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0025 | 845 | 863 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 865 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.395 | 866 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.5E-4 | 868 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1029 aa Download sequence Send to blast |
MVDRKSFVFT SPPRLDMEQL LSEAQHRWLR PAEICEILQN HHKFHIASES PTRPASGSLF 60 LFDRKVLRYF RKDGHNWRKK KDGKTIKEAH EKLKVGSIDV LHCYYAHGEG NENFQRRCYW 120 MLEQDLMHIV FVHYLEVKGN RTSIGMKENN SNSVNGTSSV NIDSTASPTS TLSSLCEDAD 180 SGDSHQASSV LRASLEPQTG NRYGWTPASG MRNISHIQRN IVRESDSQRL VDVRPWDAVG 240 NSVTRYHDQP YCNNLLTQIQ PSVTDSMLVE ENIDKGGQLK AEHFRNPLQT QSNWQIPIQD 300 DLPLPKGHVD LFSHSGMTDD TDLALFEHRA QDNFETFSSL LGSENQEPFG NSYQVPPSNM 360 ESEYIPVMKS LLRSEDSLKK VDSFSRWASK ELGEMEDLQM QSSLGDIAWT TVECETATAG 420 ITLSPSLSED QHFTIVDFWP KFAETDAEVE VMVIGTFLLS PQEVIKYNWS CMFGEVEVPA 480 EILVDGVLCC HAPPHTAGHV PFYVTCSNRF ACSELREFDF LAGSTQKIDA ADVYGPYTTA 540 SSLHLRFEKL LTHRALVHEH HIFEDVGEKR RKISKIMLLK EEKEYLLPGT YQRYSAKQEP 600 KGQLFREQFE EELYIWLIHK VTEEGKGPNM LDENGQGILH FVAALGYDWA IKPILAAGVN 660 INFRDANGWS ALHWAAFSGR EETVAVLVSL GADAGALTDP SPELPLGKTA ADLAYANGHR 720 GISGFLAESS LTSYLEKLTV DSKENSSANS SGAKAVQTVS ERTAAPMSYG DIPEKLSLKD 780 SLTAVRNATQ AADRLRQVFR MQSFQRKQLS EFGDDDKIDI SEELAVSFAA CKSKNPGHSD 840 VSLSSAATHI QKKYRGWKKR KEFLLIRQRI VKIQAHVRGH QVRKQYRTVI WSVGLLEKII 900 LRWRRKGKGL RGFKGNAIPK TVEPEQPESA ICPTKPQEDE YDFLKEGRKQ TEERLQKALT 960 RVKSMVQYPE ARDQYRRLLT VVEGFRENEA SSSASVNNEE EPVNCDEDDL IDIDSLLNDD 1020 TLMMSFSP* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
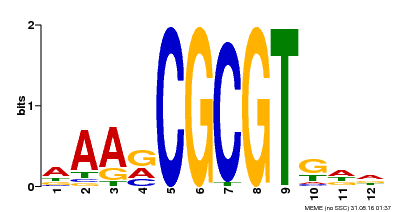
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10000120m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK228740 | 0.0 | AK228740.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 1, complete cds, clone: RAFL16-07-H14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006286969.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | R0FCP0 | 0.0 | R0FCP0_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006286969.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10000120m |
| Entrez Gene | 17884367 |



