 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10000129m | ||||||||
| Common Name | CARUB_v10000120mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1013aa MW: 114236 Da PI: 6.0711 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 186.3 | 3.3e-58 | 5 | 122 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
l+e ++rwl++ ei++iL+n++k+++++e++trp sgsl+L++rk++ryfrkDG++w+kkkdgkt++E+hekLKvg+++vl+cyYah+e n++f
Carubv10000129m 5 LSEaQHRWLRPAEICEILQNHHKFHIASESPTRPASGSLFLFDRKVLRYFRKDGHNWRKKKDGKTIKEAHEKLKVGSIDVLHCYYAHGEGNENF 98
45559***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrrcyw+Le++l +iv+vhylevk
Carubv10000129m 99 QRRCYWMLEQDLMHIVFVHYLEVK 122
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 85.154 | 1 | 127 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.4E-85 | 4 | 122 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 7.6E-51 | 7 | 121 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 5.13E-14 | 418 | 504 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 1.4E-7 | 600 | 678 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.4E-18 | 601 | 713 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.837 | 601 | 712 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.49E-19 | 602 | 712 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.90E-14 | 607 | 710 | No hit | No description |
| PROSITE profile | PS50088 | 8.656 | 618 | 650 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3500 | 618 | 647 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.22 | 651 | 683 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 651 | 680 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.51 | 826 | 848 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 6.37E-8 | 826 | 876 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.73 | 827 | 856 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0024 | 829 | 847 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 849 | 871 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.395 | 850 | 874 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.4E-4 | 852 | 871 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1013 aa Download sequence Send to blast |
MEQLLSEAQH RWLRPAEICE ILQNHHKFHI ASESPTRPAS GSLFLFDRKV LRYFRKDGHN 60 WRKKKDGKTI KEAHEKLKVG SIDVLHCYYA HGEGNENFQR RCYWMLEQDL MHIVFVHYLE 120 VKGNRTSIGM KENNSNSVNG TSSVNIDSTA SPTSTLSSLC EDADSGDSHQ ASSVLRASLE 180 PQTGNRYGWT PASGMRNISH IQRNIVRESD SQRLVDVRPW DAVGNSVTRY HDQPYCNNLL 240 TQIQPSVTDS MLVEENIDKG GQLKAEHFRN PLQTQSNWQI PIQDDLPLPK GHVDLFSHSG 300 MTDDTDLALF EHRAQDNFET FSSLLGSENQ EPFGNSYQVP PSNMESEYIP VMKSLLRSED 360 SLKKVDSFSR WASKELGEME DLQMQSSLGD IAWTTVECET ATAGITLSPS LSEDQHFTIV 420 DFWPKFAETD AEVEVMVIGT FLLSPQEVIK YNWSCMFGEV EVPAEILVDG VLCCHAPPHT 480 AGHVPFYVTC SNRFACSELR EFDFLAGSTQ KIDAADVYGP YTTASSLHLR FEKLLTHRAL 540 VHEHHIFEDV GEKRRKISKI MLLKEEKEYL LPGTYQRYSA KQEPKGQLFR EQFEEELYIW 600 LIHKVTEEGK GPNMLDENGQ GILHFVAALG YDWAIKPILA AGVNINFRDA NGWSALHWAA 660 FSGREETVAV LVSLGADAGA LTDPSPELPL GKTAADLAYA NGHRGISGFL AESSLTSYLE 720 KLTVDSKENS SANSSGAKAV QTVSERTAAP MSYGDIPEKL SLKDSLTAVR NATQAADRLR 780 QVFRMQSFQR KQLSEFGDDD KIDISEELAV SFAACKSKNP GHSDVSLSSA ATHIQKKYRG 840 WKKRKEFLLI RQRIVKIQAH VRGHQVRKQY RTVIWSVGLL EKIILRWRRK GKGLRGFKGN 900 AIPKTVEPEQ PESAICPTKP QEDEYDFLKE GRKQTEERLQ KALTRVKSMV QYPEARDQYR 960 RLLTVVEGFR ENEASSSASV NNEEEPVNCD EDDLIDIDSL LNDDTLMMSF SP* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
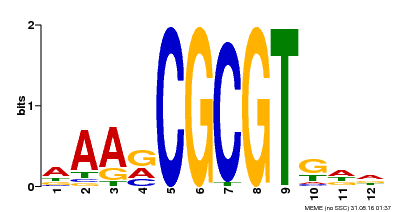
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10000129m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK228740 | 0.0 | AK228740.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 1, complete cds, clone: RAFL16-07-H14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006286969.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | R0FCP0 | 0.0 | R0FCP0_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006286969.1 | 0.0 | (Capsella rubella) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10000129m |
| Entrez Gene | 17884367 |



