 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10000560m | ||||||||
| Common Name | CARUB_v10000560mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 582aa MW: 64937.8 Da PI: 6.885 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 31.5 | 4.5e-10 | 268 | 324 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + rk + g ++ +++Aa+a++ a++k+++
Carubv10000560m 268 SIYRGVTRHRWTGRYEAHLWDnSCR-RegqARKGRQ-GGYDKEDKAARAYDLAALKYWN 324
57*******************4444.2344336655.7799999*************97 PP
| |||||||
| 2 | AP2 | 49.6 | 9.6e-16 | 367 | 418 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
Carubv10000560m 367 SIYRGVTRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 418
57****************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 7.7E-8 | 268 | 324 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.57E-13 | 268 | 333 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 2.77E-17 | 268 | 334 | No hit | No description |
| PROSITE profile | PS51032 | 16.753 | 269 | 332 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.3E-10 | 269 | 332 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.8E-18 | 269 | 338 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.3E-6 | 270 | 281 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.0E-10 | 367 | 418 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.41E-11 | 367 | 426 | No hit | No description |
| SuperFamily | SSF54171 | 1.11E-17 | 367 | 427 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 19.111 | 368 | 426 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.5E-18 | 368 | 426 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 7.9E-31 | 368 | 432 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.3E-6 | 408 | 428 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010080 | Biological Process | regulation of floral meristem growth | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0035265 | Biological Process | organ growth | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 582 aa Download sequence Send to blast |
MEMLRSSDQS QFVSYDASSA ASSSPYLLDN FYGWTNQKPQ EFFKEEAQLA AASMADSTIL 60 TTFVDPQSHH SQSHIPKLED FLGDSSSIVR YSDNSQTETQ DSSLTQIYDP RHHHHPNHNQ 120 NHHNQTGFYS DHHDFKTMAG FQTAFSTNSG SEVDDSASIG RTHLAGEYLG HVVESSGPEL 180 GFHGGSNTAN GGALSLGVNV NSSNHRNEND SNQVTTNHHY RGVNNGDRNS NNNESEKTDS 240 EKEKPIVAVE TSDCSNKKIA DTFGQRTSIY RGVTRHRWTG RYEAHLWDNS CRREGQARKG 300 RQGGYDKEDK AARAYDLAAL KYWNATATTN FPITNYSKEL EEMKHMTKQE FIASLRRKSS 360 GFSRGASIYR GVTRHHQQGR WQARIGRVAG NKDLYLGTFA TEEEAAEAYD IAAIKFRGIN 420 AVTNFEMNRY DVEAIMKSAL PIGGAAKRLK LSLEAASSEQ KPILGHQLHH FQQQQQQQQQ 480 QQLQLQSSPN HSSINFALCP NSAVQSQQII PCGIPFEAAA LYHHQQQQQQ QQHQQQNFFQ 540 HFPANAASDS TGSNNNSNVQ GSMGLMAPNP AEFFLWPNPS Y* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00504 | DAP | Transfer from AT5G10510 | Download |
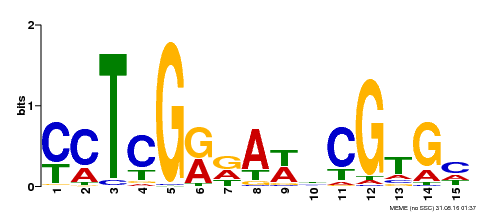
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10000560m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY974197 | 0.0 | AY974197.1 Arabidopsis thaliana clone At5g10510 AP2/EREBP transcription factor mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023635960.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL6 | ||||
| Swissprot | Q52QU2 | 0.0 | AIL6_ARATH; AP2-like ethylene-responsive transcription factor AIL6 | ||||
| TrEMBL | R0H642 | 0.0 | R0H642_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006287362.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3333 | 27 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G10510.2 | 0.0 | AINTEGUMENTA-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10000560m |
| Entrez Gene | 17881617 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



