 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10004178m | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 786aa MW: 90772 Da PI: 6.4401 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 89.9 | 3.5e-28 | 66 | 153 | 2 | 91 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHelap 91
fY+eYAk++GF++++++s++sk+++e+++++f Cs++g + e+++ +++r++++++t+Cka+++vk++ dgkw ++++ ++HnHel p
Carubv10004178m 66 FYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYGVTPESESG--SSSRRSSVKKTDCKASMHVKRRADGKWIIHEFIKDHNHELLP 153
9**************************************9999888..8999999********************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 7.8E-26 | 66 | 153 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 8.6E-26 | 274 | 367 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 8.988 | 554 | 590 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 2.2E-7 | 554 | 589 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 3.1E-7 | 565 | 592 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 786 aa Download sequence Send to blast |
MDLQENQVSD AGDECMVGIM VESNSNRDVG IVDDLKAGGD LGFSGVLDLE PRNGIDFESN 60 DAAYLFYQEY AKSMGFTTSI KNSRRSKKTK EFIDAKFACS RYGVTPESES GSSSRRSSVK 120 KTDCKASMHV KRRADGKWII HEFIKDHNHE LLPALAYHFR IQRNVKLAEK NNIDILHAVS 180 ERTRKMYVEM SRQSGGYKNI GPLLQNDDNN HVDKGRYLAL EEGDSQVLLE YFKRIKKENP 240 KFFYAIDLNE EQRLRNLFWV DAKSRDDYLS FSDVVSFDTT YVKFNDKLPL ALFIGVNHHS 300 QPMLLGCALV ADESNETFVW LIKTWLRAMG GRAPNVILTD QDKFLMAAVS ELLPNTRHCF 360 ALWHVLEKIP EYFSHVMKRH ENFLQKFNKC IFRSWTDDEF DMRWWKMVSQ FGLENDEWLL 420 WLHEHRQKWV PTFMSDVFLA GMSTSQRSES VNAFFDKYVH KKITLKEFLR QYGAILQNRY 480 EDESVADFDT CHKQPALKSP SPWEKQMATT YTHTIFKKFQ VEVLGVVACH PRKEKEDENM 540 ETFRVQDCEK DDYFLVTWSK SKSELCCFCR MFEYKGFLCR HALMILQMCG FASIPPRYIL 600 KRWTKDAKSG VMAGEGADQI QTRVQRYNDL CSRAIELSEE GCVSEENYSI VLRTLVETLK 660 NCVDLNNGQQ EADQMLESQQ SLQPMETMSS EGMSMNGYYG PQQNVQGLLN LMEPPHEGYY 720 VNQRTIQGLG QLNSIAPVQD SFFTNQQAMP GLGQMDFRPP PNFTYSLQEE HLRSAQLPGS 780 SSRQL* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
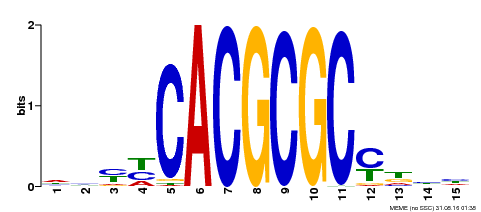
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10004178m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF159587 | 0.0 | AF159587.1 Arabidopsis thaliana far-red impaired response protein (FAR1) mRNA, complete cds. | |||
| GenBank | CP002687 | 0.0 | CP002687.1 Arabidopsis thaliana chromosome 4 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006286331.2 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A178UYQ0 | 0.0 | A0A178UYQ0_ARATH; FAR1 | ||||
| STRING | Cagra.1850s0019.1.p | 0.0 | (Capsella grandiflora) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM118 | 26 | 253 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10004178m |



