 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10008886m | ||||||||
| Common Name | CARUB_v10008706mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 512aa MW: 57471.9 Da PI: 4.6663 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 92.6 | 6.4e-29 | 3 | 78 | 52 | 128 |
NAM 52 kaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
k+++++w+fF++r++ky++ r++r t +gyWkatgkd+ + +++ vglkktLvfy+grap+ge+tdWvmhey++
Carubv10008886m 3 KTGDRQWFFFTPRNRKYPNAARSSRGTVTGYWKATGKDRVIEY-NSRSVGLKKTLVFYRGRAPNGERTDWVMHEYTM 78
56899************************************99.999****************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 36.017 | 1 | 101 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 1.83E-32 | 3 | 101 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.6E-13 | 9 | 77 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900057 | Biological Process | positive regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 512 aa Download sequence Send to blast |
MLKTGDRQWF FFTPRNRKYP NAARSSRGTV TGYWKATGKD RVIEYNSRSV GLKKTLVFYR 60 GRAPNGERTD WVMHEYTMDE EELGRCKNAK EYFALYKLYK KSGAGPKNGE QYGAPFQEEE 120 WVDSDNEEAD NVAVPDNPVV HYENTRRMDD TKFCNPISVR LEDIEKLLNE IPDAPEVTTR 180 QFNEFSGVPQ GNSVEEIQST LLNGSSGEFI DPRKTGVFLP NAQPYNRNSS FQSRLKSANS 240 FEATSGMTPL LDFEKEDYIE MDDLLIPELG ASSTEKSGQF LNNGELGDIN EFDQLFHDIS 300 MSLDVDPVFQ GTSTDMSSLS NFANNTSDQK QQFLYPQLDQ TLEYQQNNFM HPSTTLNQFT 360 DNMWFQDGQA ALYDQQPQSS SGVFTSHSAG VMPLESMNPT MSVNAQNTEG QNGGKTTSQF 420 SSALWELLES IPSTPASACE GPLNQTFVRM SSFSRIRFNG TTSVTSRKVT VAKKRISNRG 480 FLLLSIMGAL CAIFWMFIAT VGVMERPVVL S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 3e-26 | 5 | 107 | 69 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 3e-26 | 5 | 107 | 69 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 3e-26 | 5 | 107 | 69 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 3e-26 | 5 | 107 | 69 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 3e-26 | 5 | 107 | 72 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 3e-26 | 5 | 107 | 69 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 3e-26 | 5 | 107 | 69 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP) (By similarity). Transcriptional activator that promotes leaf senescence by up-regulating senescence-associated genes in response to developmental and stress-induced senescence signals. Functions in salt and oxidative stress-responsive signaling pathways. Binds to the promoter of NAC029/NAP and NAC059/ORS1 genes (PubMed:23926065). {ECO:0000250|UniProtKB:Q949N0, ECO:0000269|PubMed:23926065}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00178 | DAP | Transfer from AT1G34180 | Download |
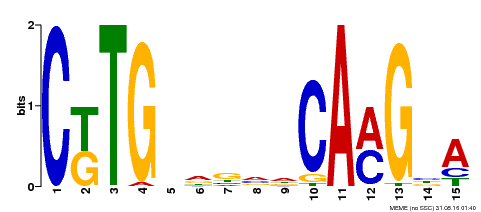
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10008886m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold, salt, drought stress and methyl methanesulfonate (MMS) treatment. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006307120.1 | 0.0 | NAC domain-containing protein 16 | ||||
| Swissprot | A4FVP6 | 0.0 | NAC16_ARATH; NAC domain-containing protein 16 | ||||
| TrEMBL | R0GVS7 | 0.0 | R0GVS7_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006307120.1 | 0.0 | (Capsella rubella) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34180.1 | 0.0 | NAC domain containing protein 16 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10008886m |
| Entrez Gene | 17900534 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



