 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10009520m | ||||||||
| Common Name | CARUB_v10009520mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 365aa MW: 39351.6 Da PI: 9.1761 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 207.1 | 5e-64 | 49 | 188 | 2 | 144 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
+++rkp+w+ErEnn+rRERrRRa+aakiy+GLRaqGny+lpk++DnneVlkALc eAGwvve+DGttyrkg kpl ++agss++a+p ss +
Carubv10009520m 49 ATRRKPSWRERENNRRRERRRRAVAAKIYTGLRAQGNYNLPKHCDNNEVLKALCSEAGWVVEEDGTTYRKGHKPL-PGDMAGSSSRATPYSSHN 141
689************************************************************************.****************** PP
DUF822 96 sslkssalaspvesysaspksssfpspssldsislasaasllpvlsvls 144
+s+ ss++ sp+ sy+ sp+sssfpsps++ ++ ++++p+l++ +
Carubv10009520m 142 QSPLSSTFDSPILSYQVSPSSSSFPSPSRVGDPHNI--STIFPFLRNGG 188
*****************************8776655..67888887765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 3.5E-61 | 50 | 171 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 365 aa Download sequence Send to blast |
LTSHSSSPSS SSSADSSSVP TGVLRICWEG KMTSDGATST SAAAAAAMAT RRKPSWRERE 60 NNRRRERRRR AVAAKIYTGL RAQGNYNLPK HCDNNEVLKA LCSEAGWVVE EDGTTYRKGH 120 KPLPGDMAGS SSRATPYSSH NQSPLSSTFD SPILSYQVSP SSSSFPSPSR VGDPHNISTI 180 FPFLRNGGIP SSLPPLRISN SCPVTPPVSS PTSRNPKPLP TWESFTKQSM SMAAKQSMTS 240 LNYPFYAVSA PASPTHHRQF HAPATIPECD ESDSSTVDSG HWISFQKFSQ QPFHGASVVP 300 ASPTFNLVKP APQQLSPNTA ATQEIGQSSD FKFENSQVKP WEGERIHDVA MEDLELTLGN 360 GKSS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 1e-35 | 51 | 133 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 1e-35 | 51 | 133 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 1e-35 | 51 | 133 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 1e-35 | 51 | 133 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 50 | 69 | RRKPSWRERENNRRRERRRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive regulator of brassinosteroid (BR) signaling. Transcription factor that activates target gene expression by binding specifically to the DNA sequence 5'-CANNTG-3'(E box) through its N-terminal domain. Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BIM1, BIM2 or BIM3. The C-terminal domain is probably involved in transcriptional activation (PubMed:12007405, PubMed:15680330, PubMed:18467490, PubMed:19170933). Recruits the transcription elongation factor IWS1 to control BR-regulated gene expression (PubMed:20139304). Forms a trimeric complex with IWS1 and ASHH2/SDG8 to regulate BR-regulated gene expression (PubMed:24838002). Promotes quiescent center (QC) self-renewal by cell divisions in the primary root. Binds to the E-boxes of the BRAVO promoter to repress its expression (PubMed:24981610). {ECO:0000269|PubMed:12007405, ECO:0000269|PubMed:15680330, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:19170933, ECO:0000269|PubMed:20139304, ECO:0000269|PubMed:24838002, ECO:0000269|PubMed:24981610}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00073 | ChIP-chip | Transfer from AT1G19350 | Download |
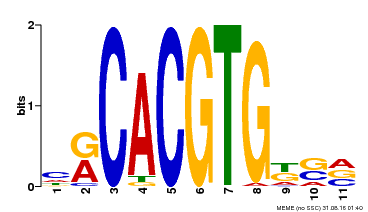
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10009520m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY065041 | 0.0 | AY065041.1 Arabidopsis thaliana At1g19350/F18O14_4 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006305153.2 | 0.0 | protein BRASSINAZOLE-RESISTANT 2 | ||||
| Refseq | XP_023644976.1 | 0.0 | protein BRASSINAZOLE-RESISTANT 2 | ||||
| Swissprot | Q9LN63 | 0.0 | BZR2_ARATH; Protein BRASSINAZOLE-RESISTANT 2 | ||||
| TrEMBL | R0GQL6 | 0.0 | R0GQL6_9BRAS; Uncharacterized protein (Fragment) | ||||
| STRING | XP_006305153.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2472 | 27 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19350.3 | 0.0 | BES1 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10009520m |
| Entrez Gene | 17900034 |



