 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10017199m | ||||||||
| Common Name | CARUB_v10017199mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 461aa MW: 51273.6 Da PI: 5.7912 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 163.1 | 1e-50 | 13 | 141 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkev 92
lp+G+rF+Ptdeel+ +yL+kk++g++ ++ ++i+e+di+k+ePwdLp + +k++++ew fF++ d+ky++g+r+nrat +gyWkatgkd+++
Carubv10017199m 13 LPVGLRFRPTDEELIRYYLRKKINGHDGDV-KAIREIDICKWEPWDLPdfSVIKTKDSEWLFFCPLDRKYPNGSRQNRATVAGYWKATGKDRKI 105
799************************999.88***************6448889999************************************ PP
NAM 93 lskkgelvglkktLvfykgrapkgektdWvmheyrl 128
+++k++ +g+k+tLvf+ grapkg++t+Wv+heyr+
Carubv10017199m 106 KTSKTNIIGVKRTLVFHAGRAPKGTRTNWVIHEYRA 141
**********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.26E-56 | 7 | 164 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 54.525 | 13 | 164 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.1E-24 | 14 | 140 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009814 | Biological Process | defense response, incompatible interaction | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0070417 | Biological Process | cellular response to cold | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 461 aa Download sequence Send to blast |
MNQNLEVLSM DSLPVGLRFR PTDEELIRYY LRKKINGHDG DVKAIREIDI CKWEPWDLPD 60 FSVIKTKDSE WLFFCPLDRK YPNGSRQNRA TVAGYWKATG KDRKIKTSKT NIIGVKRTLV 120 FHAGRAPKGT RTNWVIHEYR ATENDLSGTN PGQSPFVICK LFKKQELNLA EEDSKSDEAE 180 PAVSSPTVEV SEVTKTEDVK PYDIAESSLV ISGDSHSDAC DEATTAELVD FDWYPELESL 240 DCKLFSPFYS QVQCELGSTF QPGPSNFLGN DTNSFQVQTQ YGTNEENTHI SEFLDSILQS 300 PDEDPEKHKY VLQSGFDGAT PDQIAPAYPQ GTAVRMSNDV SVTGIKIRSR QAQPSGCTND 360 YIAQGNGPRR LRLQTNLNGS NSKNPDLKAI KREAVDTVDE TMKIGCGKLM RSKNQTGFVF 420 KRITSLKCSY GGLLRAAVVA VVFLMSVCSL TVDFRASAVP * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 9e-47 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 9e-47 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 9e-47 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 9e-47 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 9e-47 | 3 | 165 | 7 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 9e-47 | 3 | 165 | 7 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP) (PubMed:20156199, PubMed:19947982). Transcriptional activator involved in response to cold stress. Mediates induction of pathogenesis-related (PR) genes independently of salicylic signaling in response to cold. Binds directly to the PR gene promoters and enhances plant resistance to pathogen infection, incorporating cold signals into pathogen resistance responses (PubMed:19947982). Plays a regulatory role in abscisic acid (ABA)-mediated drought-resistance response (PubMed:22967043). {ECO:0000269|PubMed:19947982, ECO:0000269|PubMed:20156199, ECO:0000269|PubMed:22967043}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00398 | DAP | Transfer from AT3G49530 | Download |
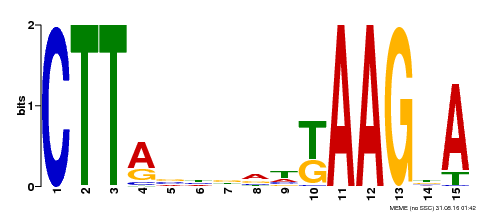
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10017199m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, drought stress, abscisic acid (ABA), salicylic acid (SA) and methyl methanesulfonate (MMS) treatment. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY059734 | 0.0 | AY059734.1 Arabidopsis thaliana putative NAC2 protein (At3g49530) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006291082.1 | 0.0 | NAC domain-containing protein 62 | ||||
| Swissprot | Q9SCK6 | 0.0 | NAC62_ARATH; NAC domain-containing protein 62 | ||||
| TrEMBL | R0HFW3 | 0.0 | R0HFW3_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006291082.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2416 | 28 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G49530.1 | 0.0 | NAC domain containing protein 62 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10017199m |
| Entrez Gene | 17886472 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



