 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10018465m | ||||||||
| Common Name | CARUB_v10018465mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 470aa MW: 52171.1 Da PI: 8.228 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 85.8 | 4.2e-27 | 277 | 329 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
r++W+ eLH++Fv+av+qL G++kA+Pk+ilelm+v+gL++e+v+SHLQk+Rl
Carubv10018465m 277 SRVVWSIELHQQFVNAVNQL-GIDKAVPKRILELMNVPGLSRENVASHLQKFRL 329
69******************.********************************8 PP
| |||||||
| 2 | Response_reg | 72.5 | 1.7e-24 | 99 | 207 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealka 92
+l+vdD+ + + +l+++l + y +v+ +++++ al+ll+e++ +Dl+l D+ mpgm+G++ll++ e +lp+i+++ g +++ ++
Carubv10018465m 99 ILVVDDDTSCLFILEKMLLRLMY-QVTICSQADVALTLLRERKdcFDLVLSDVHMPGMNGYKLLQQVGLEM-DLPVIMMSVDGRTTTVMTGINH 190
89*********************.***************99999***********************6644.8********************* PP
TESEEEESS--HHHHHH CS
Response_reg 93 GakdflsKpfdpeelvk 109
Ga d+l Kp+ peel +
Carubv10018465m 191 GACDYLIKPIRPEELKN 207
**************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF52172 | 5.41E-34 | 95 | 221 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 4.3E-40 | 96 | 220 | No hit | No description |
| SMART | SM00448 | 1.5E-28 | 97 | 209 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 41.238 | 98 | 213 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.7E-21 | 99 | 207 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.90E-26 | 100 | 212 | No hit | No description |
| PROSITE profile | PS51294 | 9.645 | 273 | 332 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.66E-18 | 274 | 333 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-27 | 274 | 334 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-23 | 277 | 329 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.4E-6 | 278 | 327 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 470 aa Download sequence Send to blast |
MAVTISASSL HDWLYTSNIA ISSLYPYSSS SSSPPTSSLT LLRAQHRNAF EAALLSAANA 60 TTGGTVNRGF HLFNTHTKAY SFLHLQMAIS DQFPTGLRIL VVDDDTSCLF ILEKMLLRLM 120 YQVTICSQAD VALTLLRERK DCFDLVLSDV HMPGMNGYKL LQQVGLEMDL PVIMMSVDGR 180 TTTVMTGINH GACDYLIKPI RPEELKNIWQ HVVRRKCTMN KELGNSQVLE DNKNSGSVET 240 VYSASECSEG SLMKRKKKKR SVDREDNEDD ENSKKSSRVV WSIELHQQFV NAVNQLGIDK 300 AVPKRILELM NVPGLSRENV ASHLQKFRLY LKRLSGVASQ SKDTECIKRY ENIQAMVSSG 360 QLHPQTLAAL FGQPVENHLS ASFGVWIPND DLGSGRSHIQ NFAVDVSSTS DRPVSVTTSM 420 AVHGLSSSAN FSQKGDADNS NRDHRIRQGY GSNVNEESWI LGKCSRQRH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-19 | 273 | 335 | 2 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 253 | 259 | KRKKKKR |
| 2 | 254 | 259 | RKKKKR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00098 | PBM | Transfer from AT2G01760 | Download |
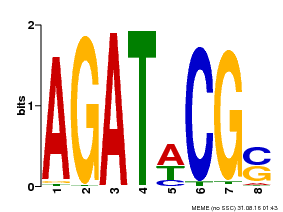
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10018465m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY088149 | 0.0 | AY088149.1 Arabidopsis thaliana clone 41875 mRNA, complete sequence. | |||
| GenBank | AY099726 | 0.0 | AY099726.1 Arabidopsis thaliana putative two-component response regulator protein (At2g01760) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023638473.1 | 0.0 | two-component response regulator ARR14 isoform X1 | ||||
| Swissprot | Q8L9Y3 | 0.0 | ARR14_ARATH; Two-component response regulator ARR14 | ||||
| TrEMBL | R0H786 | 0.0 | R0H786_9BRAS; Two-component response regulator | ||||
| STRING | XP_006292256.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8150 | 27 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01760.1 | 0.0 | response regulator 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10018465m |
| Entrez Gene | 17885868 |



