 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10020636m | ||||||||
| Common Name | CARUB_v10020636mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 330aa MW: 36492.5 Da PI: 9.7143 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 33.9 | 7.3e-11 | 39 | 90 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT eE+e l+ +++++G+g Wk I r +R+ ++k++w+++
Carubv10020636m 39 KLKWTGEEEEALLAGIAKHGPGKWKNILRDPEfadqlIHRSNIDLKDKWRNL 90
689*********************************999***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 9.806 | 34 | 95 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.87E-15 | 37 | 94 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.8E-8 | 38 | 93 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-13 | 38 | 91 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.1E-7 | 39 | 90 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 7.21E-20 | 41 | 90 | No hit | No description |
| SuperFamily | SSF46785 | 1.9E-13 | 156 | 245 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00526 | 4.9E-16 | 157 | 222 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene3D | G3DSA:1.10.10.10 | 2.5E-9 | 158 | 233 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PROSITE profile | PS51504 | 22.108 | 159 | 233 | IPR005818 | Linker histone H1/H5, domain H15 |
| Pfam | PF00538 | 9.2E-8 | 161 | 230 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 330 aa Download sequence Send to blast |
ARTNETSGDK SASDSDRAYA SRRREHKVKL HKAVMGNQKL KWTGEEEEAL LAGIAKHGPG 60 KWKNILRDPE FADQLIHRSN IDLKDKWRNL SVPPGNQGSN YKARPIKVKE EGVTPPPVTI 120 VDNINPPPIV KPIIPPVPSI QRNDLINDES FNIVVDTKNA PRYDGMIFEA LSSLADANGS 180 DVSSIFHFIE PRHEVPPNFR RILSSRLRRL AAQSKLVKVS TFKSLQNFYK IPDPSGTRTP 240 APKPKETHVK PRQSNNQASA ISQQMIDEAS ITAACKVVEA ENKIDIARVA GEEFAKITKL 300 AEGSEHLLVI ANEMHEQCSR GETVVLACV* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00231 | DAP | Transfer from AT1G72740 | Download |
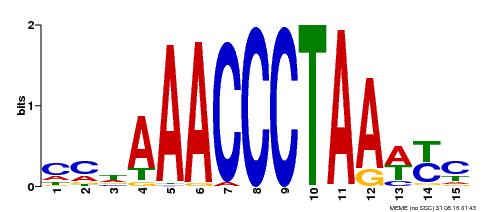
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10020636m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF372925 | 2e-75 | AF372925.1 Arabidopsis thaliana At1g72740/F28P22_7 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006302536.2 | 0.0 | telomere repeat-binding factor 5 isoform X1 | ||||
| Swissprot | F4IEY4 | 1e-148 | TRB5_ARATH; Telomere repeat-binding factor 5 | ||||
| TrEMBL | R0HZW4 | 0.0 | R0HZW4_9BRAS; Uncharacterized protein (Fragment) | ||||
| STRING | XP_006302536.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2321 | 28 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72740.1 | 1e-125 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10020636m |
| Entrez Gene | 17896538 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



