 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10023876m | ||||||||
| Common Name | CARUB_v10023876mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 261aa MW: 29380 Da PI: 8.6887 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 94.3 | 8.7e-30 | 134 | 193 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK ++++ pr+Y+rC+++ +C vkkkv+rsaedp++v+ tYeg+Hnh+
Carubv10023876m 134 VKDGYQWRKYGQKITRDNPSPRAYFRCSFSpSCLVKKKVQRSAEDPSFVVATYEGTHNHP 193
58***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.9E-28 | 125 | 195 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.83E-25 | 131 | 195 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.9E-33 | 134 | 194 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.4E-24 | 135 | 193 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 26.923 | 136 | 195 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0002237 | Biological Process | response to molecule of bacterial origin | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0031347 | Biological Process | regulation of defense response | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 261 aa Download sequence Send to blast |
MDYDLNTNPF DLHFSGKLPK REPSDSGSED VMRKWFVKDE TCMLQEELTR VNSENKKLTE 60 TLARVSEKYS ELQNLFKELQ SRENVNFCNE RKRKQDRDEF VSSPIGISCG GTVEKSTVST 120 AYFHAEKSDT SLTVKDGYQW RKYGQKITRD NPSPRAYFRC SFSPSCLVKK KVQRSAEDPS 180 FVVATYEGTH NHPGPNASVS RTVKLDLVQG GLEPVEEKKG RGTIQDVLVQ QMASSLTKDP 240 KFTAALAAAI SGRFIEQSRT * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 4e-20 | 136 | 192 | 19 | 74 | Probable WRKY transcription factor 4 |
| 2lex_A | 4e-20 | 136 | 192 | 19 | 74 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00068 | PBM | Transfer from AT2G25000 | Download |
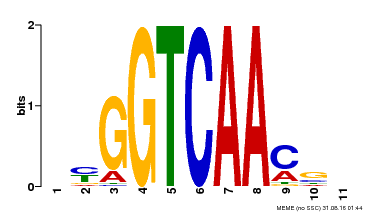
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10023876m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK227987 | 1e-171 | AK227987.1 Arabidopsis thaliana mRNA for putative WRKY-type DNA binding protein, complete cds, clone: RAFL14-52-F05. | |||
| GenBank | BT004611 | 1e-171 | BT004611.1 Arabidopsis thaliana At2g25000 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006294825.1 | 0.0 | probable WRKY transcription factor 60 isoform X1 | ||||
| Swissprot | Q9SK33 | 1e-151 | WRK60_ARATH; Probable WRKY transcription factor 60 | ||||
| TrEMBL | R0HUF2 | 0.0 | R0HUF2_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006294825.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6584 | 19 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G25000.1 | 1e-153 | WRKY DNA-binding protein 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10023876m |
| Entrez Gene | 17889950 |



