 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10025787m | ||||||||
| Common Name | CARUB_v10025787mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1056aa MW: 117812 Da PI: 6.365 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 188.3 | 7.8e-59 | 19 | 136 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
l+e ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+e+n++f
Carubv10025787m 19 LSEaQHRWLRPAEICEILRNHQKFHIASEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEDNENF 112
45559***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrrcyw+Le++l +iv+vhylevk
Carubv10025787m 113 QRRCYWMLEQDLMHIVFVHYLEVK 136
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 86.275 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.6E-87 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.4E-52 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.06E-13 | 465 | 549 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 7.0E-19 | 646 | 757 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.28 | 646 | 757 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.7E-18 | 647 | 758 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.3E-7 | 651 | 723 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.19E-14 | 652 | 755 | No hit | No description |
| SMART | SM00248 | 3200 | 663 | 692 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.71 | 663 | 695 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 696 | 725 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 10.873 | 696 | 728 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.32 | 871 | 893 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 8.23E-8 | 872 | 921 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.002 | 873 | 892 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.858 | 873 | 901 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 894 | 916 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.304 | 895 | 919 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.6E-4 | 897 | 916 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1056 aa Download sequence Send to blast |
MADRGSFGFA PRLDIKQLLS EAQHRWLRPA EICEILRNHQ KFHIASEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEDNE NFQRRCYWML 120 EQDLMHIVFV HYLEVKGNRM STGGTKENHS NSLSGTGSVN VDSTATRSSI LSPLCEDADS 180 GDSHQASSSL QPNPEPQIMH HQNASTMNSF NTSSVLGNRD GWTSAPAIGI VPQVHENRVK 240 ESDSQRSGDV PVWDASFENS LARYQNLPYN APSTQTQPSN FGLMPMEGKT GSLLTAEHLR 300 NPLQNQVNWQ IPVQESLPLQ KWPMDSHSGM TDATDLALFG QRAHGNFGTF SSLLGCQNQQ 360 PSSFQAPFTN NEAAYIPKLG PEDLIYEASA NQTLPLRKAL LKTEDSLKKV DSFSRWVSKE 420 LGEMEDLQMQ SSSGGIAWTS VECETAAAGS SLSPSLSEVQ RFTMIDFWPK WTQTDSEVEV 480 MVIGTFLLSP KEVTSYSWSC MFGEVEVPAE ILVDGVLCCH APPHEVGRVP FYITCSDRFS 540 CSEVREFDFL PGSTRKLNAT DVYGANTIEA SLHLRFENLL ALRSSVQEHH IFENVGQKRR 600 KISKIMLLMD EKESLLPGTI EKDSTELEAK ERLIREEFED KLYLWLIHKV TEEGKGPNIL 660 DEVGQGVLHL AAALGYDWAI KPILAAGVSI NFRDANGWSA LHWAAFSGRE DTVAVLVSLG 720 ADAGALADPS PEHPLGKTAA DLAYGNGHRG ISGFLAESSL TSYLEKLTVD AKENSSADSS 780 GAKAVLTVAE RTATPMSYGD VPETLSMKDS LTAVLNATQA ADRLHQVFRM QSFQRKQLSE 840 LGGDNEFDIS DELAVSFAAA KTKKPGHSNG AVHAAAVQIQ KKYRGWKKRK EFLLIRQRIV 900 KIQAHVRGHQ VRKQYRAIIW SVGLLEKIIL RWRRKGSGLR GFKRDTITKP PEPVCPAPQE 960 DDYDFLKEGR KQTEERLQKA LTRVKSMAQY PEARAQYRRL LTVVEGFREN EASSSSALRN 1020 NNSNTEEAAN YNEEDDLIDI DSLLDDDTFM SLTFE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
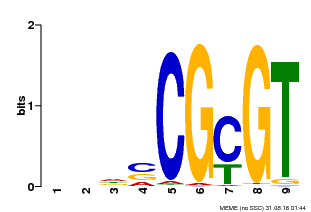
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10025787m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB493811 | 0.0 | AB493811.1 Arabidopsis thaliana At5g64220 mRNA for hypothetical protein, partial cds, clone: RAAt5g64220. | |||
| GenBank | AK229403 | 0.0 | AK229403.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 2, complete cds, clone: RAFL16-66-C08. | |||
| GenBank | BT010874 | 0.0 | BT010874.1 Arabidopsis thaliana At5g64220 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006279929.1 | 0.0 | calmodulin-binding transcription activator 2 isoform X1 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | R0EUM4 | 0.0 | R0EUM4_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006279929.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10025787m |
| Entrez Gene | 17876934 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



