 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10026295m | ||||||||
| Common Name | CARUB_v10026295mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 490aa MW: 54272.7 Da PI: 6.6815 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 33.2 | 1.3e-10 | 230 | 287 | 1 | 56 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pseng...krkrfslgkfgtaeeAakaaiaarkklege 56
s y+GV++++++gr++A+++d + rk + g ++ +++Aa+a++ a++k++g+
Carubv10026295m 230 SIYRGVTRHRWTGRYEAHLWDnSCR-RegqARKGRQ-GGYDKEDKAARAYDLAALKYWGH 287
57*******************4444.2344336655.7799999*************997 PP
| |||||||
| 2 | AP2 | 50.8 | 4.2e-16 | 329 | 380 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
Carubv10026295m 329 SVYRGVTRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 380
78****************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.6E-8 | 230 | 287 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.63E-18 | 230 | 296 | No hit | No description |
| SuperFamily | SSF54171 | 5.1E-14 | 230 | 295 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 4.8E-20 | 231 | 300 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.214 | 231 | 294 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.8E-11 | 231 | 295 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-6 | 232 | 243 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.1E-10 | 329 | 380 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 8.43E-11 | 329 | 388 | No hit | No description |
| SuperFamily | SSF54171 | 9.15E-18 | 329 | 389 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 6.6E-30 | 330 | 394 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.137 | 330 | 388 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.0E-18 | 330 | 388 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-6 | 370 | 390 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 490 aa Download sequence Send to blast |
MAPPPMTNWL TFSRSPMELL KSPEHSHFPS SFDASSTPFL LDNFYEEAEI EAAATMADST 60 TLSTYFDSQI HSQTQIPKLE DFLGDSFVRY SDNQTETQDS SSSLTRFYDL SHHTVTGGVT 120 GFFSDHHDFK AVNSGSEIVD DSASNIVGTH LSGHAVVKSS STTAELGFNG GATTGEALSL 180 AVNNTSDQPL SCDNGERREN SKKMKTTVSK KETSDDSKKK VVEPLGQRTS IYRGVTRHRW 240 TGRYEAHLWD NSCRREGQAR KGRQGGYDKE DKAARAYDLA ALKYWGHTAT TNFPAASYSK 300 EVEDMNHMTK QEFIASLRRK SSGFSRGASV YRGVTRHHQQ GRWQARIGRV AGNKDLYLGT 360 FATEEEAAEA YDIAAIKFRG INAVTNFEMN RYDVEAVMKS SFPVGNSAKR HKLAVESPPS 420 SDHNLQQPLL PSSSSSDVNP NSIPCGIPFE SSVLYHHQNF FQHYPLASDS TVQAPMNPAE 480 FFLWTNPSY* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00589 | DAP | Transfer from AT5G65510 | Download |
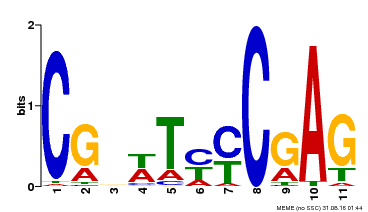
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10026295m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY560887 | 0.0 | AY560887.1 Arabidopsis thaliana putative AP2/EREBP transcription factor (At5g65510) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023636767.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL7 isoform X1 | ||||
| Refseq | XP_023636770.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL7 isoform X1 | ||||
| Swissprot | Q6J9N8 | 0.0 | AIL7_ARATH; AP2-like ethylene-responsive transcription factor AIL7 | ||||
| TrEMBL | R0EWM6 | 0.0 | R0EWM6_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006280368.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3333 | 27 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65510.1 | 0.0 | AINTEGUMENTA-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10026295m |
| Entrez Gene | 17874929 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



