 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Carubv10028472m | ||||||||
| Common Name | CARUB_v10028472mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Capsella
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 279aa MW: 31562.2 Da PI: 4.7425 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 66.5 | 5e-21 | 134 | 184 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+++AeIrdp+++g r++lg+f+ta eAa+a+++a+ +l+g
Carubv10028472m 134 HYRGVRQRP-WGKFAAEIRDPNKRG--SRVWLGTFDTAIEAARAYDQAAFRLRG 184
69*******.**********96655..*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.49E-31 | 133 | 192 | No hit | No description |
| Pfam | PF00847 | 4.1E-15 | 134 | 184 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.421 | 134 | 192 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.1E-33 | 134 | 193 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-23 | 134 | 194 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 2.7E-37 | 134 | 198 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.5E-11 | 135 | 146 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.5E-11 | 158 | 174 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 279 aa Download sequence Send to blast |
MATPNEVSAL WFIEKHLLDD LSPVATTHEP PPASESSSDS CFSDSQIIDL DTPRFVDFIL 60 HSDISLSDFD FKPTHQNQYQ NQNQVEPEIN NSQIRKPPLK IALPAKTEWI QFVAEDLNPK 120 PEVTKPEVEE KKQHYRGVRQ RPWGKFAAEI RDPNKRGSRV WLGTFDTAIE AARAYDQAAF 180 RLRGSKAILN FPLEVGKWKP RADEPTENKR KRDNDGDDDD EKATLVEKAL KTEQNVDGDG 240 GETFPLYTDW DLTGFLNFPL LSPLSPHPSF GYSQLTVV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 3e-31 | 131 | 195 | 2 | 66 | ATERF1 |
| 3gcc_A | 3e-31 | 131 | 195 | 2 | 66 | ATERF1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. Involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. {ECO:0000269|PubMed:10715325, ECO:0000269|PubMed:9756931}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00549 | DAP | Transfer from AT5G47230 | Download |
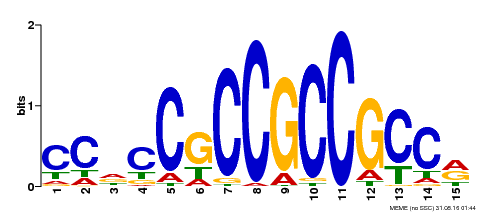
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Carubv10028472m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Ethylene induction is completely dependent on a functional ETHYLENE-INSENSITIVE2 (EIN2). Wounding as well as cold stress induction does not require EIN2. Transcripts accumulate strongly in cycloheximide-treated plants, a protein synthesis inhibitor. Seems to not be influenced by jasmonate, Alternaria brassicicola, exogenous abscisic acid (ABA), cold, heat, NaCl or drought stress. {ECO:0000269|PubMed:10715325}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006282206.1 | 0.0 | ethylene-responsive transcription factor 5 | ||||
| Swissprot | O80341 | 1e-133 | EF102_ARATH; Ethylene-responsive transcription factor 5 | ||||
| TrEMBL | R0GEF7 | 0.0 | R0GEF7_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006282206.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10 | 28 | 1650 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47230.1 | 1e-104 | ethylene responsive element binding factor 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Carubv10028472m |
| Entrez Gene | 17876679 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



