 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc01_g15290 | ||||||||
| Common Name | GSCOC_T00016581001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 488aa MW: 54002 Da PI: 6.5358 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 34.4 | 5e-11 | 438 | 485 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqky 47
rW+t E+e l +v+++G g+Wk I + Rt+ ++k++w+++
Cc01_g15290 438 RWSTSEEETLRIGVEKYGRGNWKVILNAYRdvfDERTEVDLKDKWRNL 485
9********************************99***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-11 | 429 | 486 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.184 | 431 | 487 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.43E-14 | 433 | 486 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.5E-4 | 435 | 487 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 1.99E-18 | 437 | 487 | No hit | No description |
| Pfam | PF00249 | 8.9E-9 | 438 | 485 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 488 aa Download sequence Send to blast |
MDVDIDVDIA RWILEFLVRQ PLDDRIVSSL ISILPPTNEN SGLKKSLLLR RIESEISAGS 60 VSEKILDILE QIEELDFQDK IKQHSDLIKA AYCSVAVACT AKFLNDKDVD SKFRYFDAVR 120 RIWRGRVCKM ENAEKVGLIS EELKSWKDEL EAAVWDESIW DSVLQKSKGL DAVGAVRLYV 180 REEKDDIGPS FLELVAEALR ADDKLKGILG FKGTHGGAIQ SSGPRDANTG GCFSIHVEVQ 240 RANVVSGRKH VASKRIRGAP SGMSRGAKII DGDTSGADIS GKTFDLPSTP EVHEVQEALK 300 SSSLDLKAVV KDPLPDALNI ADTVVSSIER KDAGKQPVEE NHVGANPSIA ESSGAVQANE 360 GTLGNHCSEH QKDAPRQSLM ARSSTARTFE WDDSVDALAG DSPNPGNGVQ LPSPKRRVVS 420 PLKKYEINNM KKRRKFKRWS TSEEETLRIG VEKYGRGNWK VILNAYRDVF DERTEVDLKD 480 KWRNLTR* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 414 | 434 | KRRVVSPLKKYEINNMKKRRK |
| 2 | 430 | 434 | KKRRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
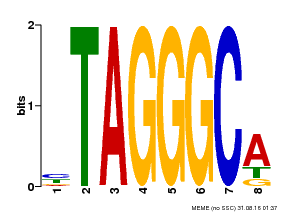
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027091281.1 | 0.0 | uncharacterized protein LOC113712180 isoform X1 | ||||
| Refseq | XP_027112453.1 | 0.0 | uncharacterized protein LOC113731391 isoform X1 | ||||
| TrEMBL | A0A068U6V9 | 0.0 | A0A068U6V9_COFCA; Uncharacterized protein | ||||
| STRING | XP_009587848.1 | 1e-153 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3774 | 24 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 4e-14 | TRF-like 5 | ||||



