 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc01_g17380 | ||||||||
| Common Name | GSCOC_T00016331001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 996aa MW: 110468 Da PI: 5.0852 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 59.1 | 7.6e-19 | 854 | 950 | 2 | 95 |
EEE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEE.TTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS.SEE..E CS
B3 2 fkvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltled.esgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFkldgr.sefelv 93
+kvl++sdv+++gr+vlpkk ae+h +++++ ++ + led ++++W++++++ ++ks++y+l+ ++ +Fv+ ngL+egDf+v++ ++ ++++++
Cc01_g17380 854 QKVLKQSDVGNLGRIVLPKKEAETHlpELESRDGIPIALEDiGTSNVWNMRYRFwpNNKSRMYLLE-NTGDFVRVNGLQEGDFIVIY--SDtKCGKYL 948
79*************************888999999*****7778*******99777777777777.********************..444898887 PP
EE CS
B3 94 vk 95
++
Cc01_g17380 949 IR 950
76 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 1.7E-27 | 847 | 962 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10015 | 2.61E-33 | 851 | 952 | No hit | No description |
| SuperFamily | SSF101936 | 2.09E-22 | 852 | 947 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 10.503 | 853 | 955 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 8.6E-20 | 853 | 955 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 3.8E-15 | 854 | 949 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 996 aa Download sequence Send to blast |
MKQDEQVMSV NGGCGEREIW FGRDHRGHDQ DNSLLDDDDN DADVNNDPCS LFYTDFPPIP 60 DFPCMSSSSS SSSTPLPARA VASTSSSSSS SSSSAATSWA VFKDMNCMNA MENFGYMDLL 120 DNSDIWDPCS IFESENHHQP LEEDGNDNGN DGGLYFLQAN SELGVIFLEW LKQNKDHISA 180 EDMRSIKLKR STIDCASKRL GSTKEGKKQL LKLILEWVEQ YQLHKRRATE MVVITKGTRQ 240 NSHGDGVEAD CRMSKNASEA LQAGDDGDGL IKGMDVIMEE SAFPATAFDD DEDHMKQDEQ 300 VMSVNGGCGE REIWFGRDHR GHDQDNSLLD DDDNDADVNN DPCSLFYTDF PPIPDFPCMS 360 SSSSSSSTPL PARAVASTSS SSSSSSSSAA TSWAVFKSEA DDSFHPNSEA EMQRYHPLER 420 QCDEVVNAAA PAAALSSTAS MEILPLPDDY CSNRDMNCMN AMENFGYMDL LDNSDIWDPC 480 SIFESENVNP PQQDHDHQFQ ERKPAPSAYG NDNGNDGGLY FLQANSELGV IFLEWLKQNK 540 DHISAEDMRS IKLKRSTIDC ASKRLGSTKE GKKQLLKLIL EWVEQYQLHK RRATEMDPST 600 ACLSPSPSPW VPPAPPCESC PNSLAAPFPP TMGGYARDPY SSAAAATVAV PVPVSNQTMN 660 NANLYLVPAE YHQPTDSPQS WSGPYSMPQA QYNPFPDNTK LPVSGSQAQA LYANPYPYQV 720 FDGSSGERLV RYGSSATKEA RKKRMARQRR ISLHQYRHHH THQNQPQSLV NEQHATRIPA 780 VGDDCTSNSR ANPGNWIYWS PAVPPPAPAP MASTMPPLVS SDKPSHRQAP TQPVPSSDRK 840 QSWKTEKNLK FLLQKVLKQS DVGNLGRIVL PKKEAETHLP ELESRDGIPI ALEDIGTSNV 900 WNMRYRFWPN NKSRMYLLEN TGDFVRVNGL QEGDFIVIYS DTKCGKYLIR GVKVRQPGSK 960 SEAKKPARRN QRNVPQAANN FAFASFKQTV KRTTN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9b_A | 1e-36 | 849 | 956 | 1 | 108 | B3 domain-containing transcription factor FUS3 |
| 6j9b_D | 1e-36 | 849 | 956 | 1 | 108 | B3 domain-containing transcription factor FUS3 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
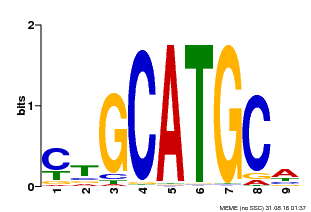
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027174476.1 | 0.0 | B3 domain-containing transcription factor ABI3 | ||||
| TrEMBL | A0A068U8A0 | 0.0 | A0A068U8A0_COFCA; Uncharacterized protein | ||||
| STRING | XP_009792996.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4270 | 23 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 3e-85 | B3 family protein | ||||



