 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc02_g16110 | ||||||||
| Common Name | GSCOC_T00014086001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 267aa MW: 30788.9 Da PI: 9.9457 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 98.7 | 2.3e-31 | 25 | 74 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krien++nrqvtf+kRrng+lKKA+ELSvLCdaeva+i+fs++g+lyey+
Cc02_g16110 25 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYA 74
79***********************************************8 PP
| |||||||
| 2 | K-box | 112 | 6.3e-37 | 94 | 189 | 5 | 100 |
K-box 5 sgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
++ s +ea+a+++qqe++kL+++i+nLq+++R++lGe+L+sL+l+eL+++e+++e++++++RskKnell+++ie +qk+e +l+++n++Lr+k++e
Cc02_g16110 94 NTGSISEANAQHYQQEASKLRAQISNLQNSNRNMLGESLGSLNLRELKNIESKVERGISRVRSKKNELLFAEIEFMQKREVDLHNNNQYLRSKIAE 189
444599***************************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 33.613 | 17 | 77 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 7.1E-41 | 17 | 76 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.88E-44 | 18 | 94 | No hit | No description |
| SuperFamily | SSF55455 | 6.28E-33 | 18 | 90 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 19 | 73 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.3E-33 | 19 | 39 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.4E-26 | 26 | 73 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.3E-33 | 39 | 54 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.3E-33 | 54 | 75 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 3.0E-26 | 100 | 187 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 15.09 | 103 | 193 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 267 aa Download sequence Send to blast |
MSCQSDPSRE TSPQRKLGRG KIEIKRIENT TNRQVTFCKR RNGLLKKAYE LSVLCDAEVA 60 LIVFSNRGRL YEYANNSVKE TIKRYKTVNS DSANTGSISE ANAQHYQQEA SKLRAQISNL 120 QNSNRNMLGE SLGSLNLREL KNIESKVERG ISRVRSKKNE LLFAEIEFMQ KREVDLHNNN 180 QYLRSKIAET ERAQHDMNLM PGSSDYELVS AQPFDARTFL QVNGLQLNNH YPRQEQRPLQ 240 LVYDFLTSLF TTHQLTYMLR PLIMHL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tqe_P | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_Q | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_R | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 1tqe_S | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 5f28_A | 3e-20 | 17 | 101 | 1 | 87 | MEF2C |
| 5f28_B | 3e-20 | 17 | 101 | 1 | 87 | MEF2C |
| 5f28_C | 3e-20 | 17 | 101 | 1 | 87 | MEF2C |
| 5f28_D | 3e-20 | 17 | 101 | 1 | 87 | MEF2C |
| 6c9l_A | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_B | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_C | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_D | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_E | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| 6c9l_F | 4e-20 | 17 | 85 | 1 | 69 | Myocyte-specific enhancer factor 2B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in regulating genes that determines stamen and carpel development in wild-type flowers. {ECO:0000250}. | |||||
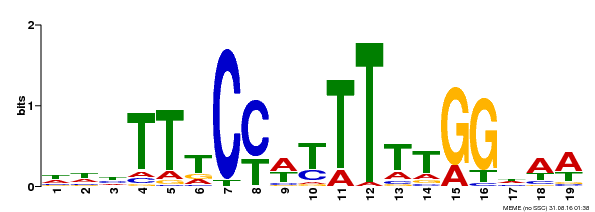
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU332281 | 0.0 | GU332281.1 Coffea arabica MADS-box protein AG subfamily (C03) mRNA, complete cds. | |||
| GenBank | KJ483236 | 0.0 | KJ483236.1 Coffea arabica AG (AG) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027112013.1 | 1e-176 | floral homeotic protein AGAMOUS-like | ||||
| Refseq | XP_027160914.1 | 1e-176 | floral homeotic protein AGAMOUS | ||||
| Swissprot | Q40885 | 1e-143 | AG_PETHY; Floral homeotic protein AGAMOUS | ||||
| TrEMBL | A0A068TLQ2 | 0.0 | A0A068TLQ2_COFCA; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400073134 | 1e-140 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 1e-117 | MIKC_MADS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



