 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc02_g31850 | ||||||||
| Common Name | GSCOC_T00027482001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 545aa MW: 61733 Da PI: 6.887 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 46.9 | 7.2e-15 | 126 | 187 | 2 | 68 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikege 68
W+++evlaL+++r++me+++ + We+vs+k+ g++rs+++Ckek+e+ ++ + ++ ++
Cc02_g31850 126 WSNDEVLALLRIRSSMENWFPEI-----TWEHVSRKLTGLGYHRSAEKCKEKFEEESRHFNSMNYNK 187
********************998.....9****************************9998877554 PP
| |||||||
| 2 | trihelix | 93.5 | 2e-29 | 430 | 519 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgk.....lkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+++ev+aLi+++ ++ +++ ++ +k+plWe++s+ m e g++rs+k+Ckekwen+nk+++k+k+++kkr s +s+tcpyf+ql
Cc02_g31850 430 RWPRDEVQALINLKCRLTNNSTSDDsikegAKGPLWERISQGMLELGYKRSSKRCKEKWENINKYFRKTKDNNKKR-SLDSRTCPYFHQLS 519
8*************99888888653344559********************************************8.9999********95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 19 | 122 | 179 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 8.8E-11 | 124 | 197 | No hit | No description |
| PROSITE profile | PS50090 | 6.365 | 125 | 177 | IPR017877 | Myb-like domain |
| PROSITE profile | PS50090 | 7.062 | 423 | 492 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.039 | 427 | 494 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 2.95E-22 | 429 | 499 | No hit | No description |
| Pfam | PF13837 | 1.8E-18 | 429 | 519 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 545 aa Download sequence Send to blast |
MFDGVPADQF HQFLAASSRT SLPIPLSFPL HHHGVSIPSS AASVSLASPP LPPTSATPPA 60 PAFLGCFDPY SSPLTLDVQV QPHHHHQQSG LHHQLHHQSP PPTSKNGEEK EEREGSISAA 120 IPLDPWSNDE VLALLRIRSS MENWFPEITW EHVSRKLTGL GYHRSAEKCK EKFEEESRHF 180 NSMNYNKNYR FFSDLDELYN DENPQVSTEK SQDLVKEKDK QEGDNKMDAS TLQEEEDTGN 240 TVAVANPSDQ ENAELVKEST KSRKRKRNHK FEMFKGFCEA VVKKIVEQQE VLHNKLIEDM 300 VRRDRESIAR DEAWKCQEMD RINKEIEMRA QEQAIACDRQ GKIIDLLKKF TSGSEADQSL 360 VRRIEDLLKV TNSSNSVTSS SEILPPSSLN SDQTKLEAVT SASMAISHQN PTLKMVSAPN 420 ERGERIAGKR WPRDEVQALI NLKCRLTNNS TSDDSIKEGA KGPLWERISQ GMLELGYKRS 480 SKRCKEKWEN INKYFRKTKD NNKKRSLDSR TCPYFHQLST LYGQGTLVAP SNLPENHQTS 540 PENH* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
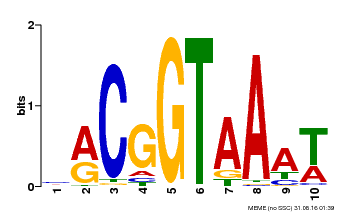
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027107424.1 | 0.0 | trihelix transcription factor GTL2-like | ||||
| TrEMBL | A0A068UJ84 | 0.0 | A0A068UJ84_COFCA; Uncharacterized protein | ||||
| STRING | VIT_14s0030g01860.t01 | 1e-154 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA13700 | 10 | 13 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 1e-92 | Trihelix family protein | ||||



