 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc02_g37380 | ||||||||
| Common Name | GSCOC_T00027998001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 831aa MW: 90045.4 Da PI: 6.4236 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.1 | 4.7e-21 | 125 | 180 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Cc02_g37380 125 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 180
688999***********************************************999 PP
| |||||||
| 2 | START | 203.7 | 7.8e-64 | 338 | 565 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevi 85
ela++a++elvk+a+ +ep+W +s e++n +e++++f++ + + +ea+r++g+v+ ++ lve+l+d + +W e+++ + +t++vi
Cc02_g37380 338 ELALAAMNELVKLAQTDEPLWLRSLeggrEILNHEEYMRTFTPCIGvkptgFVTEASRETGMVIINSLALVETLMDAN-KWAEMFPcmiaRTSTTDVI 434
5899**************************************99889*******************************.******************* PP
CTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE-- CS
START 86 ssg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlk 173
s g galqlm elq+lsplvp R++ f+R+++q+ +g+w++vdvSvds ++ + s+ + R+++lpSg+l+e+++ng+skvtwveh+d++
Cc02_g37380 435 SGGmggtrnGALQLMHGELQVLSPLVPvREVNFLRFCKQHAEGVWAVVDVSVDSIRETSGggaSPTFPRSRRLPSGCLVEDMPNGYSKVTWVEHADYD 532
*********************************************************999999*********************************** PP
SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 174 grlphwllrslvksglaegaktwvatlqrqcek 206
++++h+l+r+l++ g+ +g ++w atlqrqce+
Cc02_g37380 533 ESMIHQLYRPLIGAGMGFGSQRWIATLQRQCEC 565
*******************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.54E-20 | 109 | 182 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-21 | 112 | 182 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 122 | 182 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.9E-18 | 123 | 186 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.35E-18 | 124 | 182 | No hit | No description |
| Pfam | PF00046 | 1.4E-18 | 125 | 180 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 157 | 180 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 45.064 | 329 | 568 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.35E-33 | 331 | 565 | No hit | No description |
| CDD | cd08875 | 3.07E-121 | 333 | 564 | No hit | No description |
| SMART | SM00234 | 1.6E-53 | 338 | 565 | IPR002913 | START domain |
| Pfam | PF01852 | 4.7E-56 | 338 | 565 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 1.4E-4 | 443 | 538 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 5.44E-24 | 586 | 815 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 831 aa Download sequence Send to blast |
MNFGGFLDNN SVGGGGAKIV ADIPYSDSNN VNSSNTNMPA SAAIAQPRLA TQSLSKSMFS 60 SPGLSLALQT SLEGQEVRRM SENYESNMNF GRRSRDEEHE SRSGSDNMEG GSGDDQEAAD 120 KPPRKKRYHR HTPQQIQELE ALFKECPHPD EKQRLELSKR LCLETRQVKF WFQNRRTQMK 180 TQLERHENSI LRQENDKLRA ENMSIREAMR NPICTNCGGP AIIGEVSLEE QHLRIENARL 240 KDELDRVCTL AGKFLGRPIS SSAASMAPPL PNSSLELGVG GNGFASLSTV PATLPLGPPD 300 FGVGIGNPLS VMAGPTKAAA TTGGTGIERS LEKSMYLELA LAAMNELVKL AQTDEPLWLR 360 SLEGGREILN HEEYMRTFTP CIGVKPTGFV TEASRETGMV IINSLALVET LMDANKWAEM 420 FPCMIARTST TDVISGGMGG TRNGALQLMH GELQVLSPLV PVREVNFLRF CKQHAEGVWA 480 VVDVSVDSIR ETSGGGASPT FPRSRRLPSG CLVEDMPNGY SKVTWVEHAD YDESMIHQLY 540 RPLIGAGMGF GSQRWIATLQ RQCECLAILM SSSVAPRDHT AITASGRRSM LKLAQRMTDN 600 FCAGVCASTV HKWNKLCAGN VDEDVRVMTR KSVDDPGEPP GIVLSAATSV WLPVSPQRLF 660 DFLRDERLRS EWDILSNGGP MQEMAHIAKG QDHGNCVSLL RASAMNANQS SMLILQETCI 720 DAAGSLVVYA PVDIPAMHVV MNGGDSAYVA LLPSGFAIVP DGPGSRGPLP NGLTNGPTSH 780 RVSGSLLTVA FQILVNSLPT AKLTVESVET VNNLISCTVQ KIKAALHCES * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
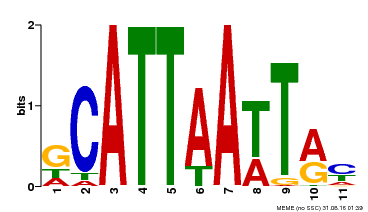
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027108261.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2-like | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A068UK79 | 0.0 | A0A068UK79_COFCA; Uncharacterized protein | ||||
| STRING | XP_009602856.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1626 | 24 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



