 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc04_g10620 | ||||||||
| Common Name | GSCOC_T00042622001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1282aa MW: 141448 Da PI: 8.5649 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.1 | 0.00029 | 1190 | 1212 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
Cc04_g10620 1190 CPvkGCGKKFFSHKYLVQHRRVH 1212
9999*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.0E-13 | 26 | 67 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.425 | 27 | 68 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 1.8E-14 | 28 | 61 | IPR003349 | JmjN domain |
| SMART | SM00558 | 2.2E-50 | 193 | 362 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.102 | 196 | 362 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 8.79E-25 | 208 | 360 | No hit | No description |
| Pfam | PF02373 | 2.9E-36 | 226 | 345 | IPR003347 | JmjC domain |
| SMART | SM00355 | 23 | 1165 | 1187 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.0045 | 1188 | 1212 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.965 | 1188 | 1217 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.3E-6 | 1190 | 1216 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1190 | 1212 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 8.35E-10 | 1204 | 1246 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 7.0E-9 | 1217 | 1242 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0016 | 1218 | 1242 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.824 | 1218 | 1247 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1220 | 1242 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 5.97E-8 | 1236 | 1270 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.2E-9 | 1243 | 1271 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.83 | 1248 | 1274 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.658 | 1248 | 1279 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1250 | 1274 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1282 aa Download sequence Send to blast |
MASEASSTPV ASGNIEVFSW LKTLPVAPEY HPTLAEFQDP IAYIFKIEKE ASQYGICKIV 60 PPVPAPHKKT AVSNLNKSLV ARSGSPTFTT RQQQIGFCPR KQRPVQKPVW QSGENYTLEE 120 FEAKAKAFER NYLKKSRKKC LIPLEIETLY WKAIVDKPFS VEYANDMPGS AFAPRRGGKE 180 GGGEGSNVNA NVTVGDTEWN MRGVSRAKGS LLTFMKEEIP GVTSPMVYVA MMFSWFAWHV 240 EDHDLHSLNY LHTGAGKTWY GVPWDAAAAF EEVIRVHGYG GEINPLVTFS TLGEKTTVMS 300 PEVLIDAGVP CCRLVQNAGE FVVTFPRAYH SGFSHGFNCG EAANIATPGW LTVAKDAAIR 360 RASINCPPMV SHFQLLYDLA LSSCSRVPRG VRMEPRSSRL KDKKKGEGEM LVKDLFVQDV 420 MQNNDLLYML AEGSSVVILP QNSVVSSFSS NSKAGSQSQV QPGLFPSLGS PDLMMKTTKS 480 LLSEGIVQER KRGVLQGTGS CSMKETVSPS CFDKRVPCSV RGNEFSALAS ESKNMETEKG 540 RASRGDRLSE QGLFSCVTCG ILCFACVAIV QPTDAAATYL ITADRSEFKD WGETSDVSTV 600 VNGDEVLPKS DSCSGWMYKR NPDELFDVPV QSGGLYQSVD DEIVGLIPNT EAQKDTSSLG 660 LLALTYGNSS DSDEDDVDAN NHTEACQNEA KDCSPESGLY CHDAGLHKGG SRNDVFSCSE 720 FSCADVVPLQ IIGSSDKQGT TKSTSESRRH PPPDGTIEYK RRSFPLMEID NLADRCRHQV 780 KEQDASSPSP LAHKAETIAS TAIVEFENKT LPFAGRPDED SSRMHVFCLQ HAVQVEKQLR 840 SIGGVNVLLL CHPDYPNVEA QAKKMAEELG GHYVWSNISF RQASKEDEET IQAALESQEA 900 IHGNGDWAVK LGINLYYSAS LSRSPLYSKQ MPYNLVIYNA FGRSSPVNSP TKDDSLGKGP 960 GKPKKTVVAG KWCGKIWMSN QVHPFLAERD EEEQERGIPS CMKADLKPDR PLESTRVQTG 1020 ETTARTCRTG RKRKAAAEIR PAVKAKSAKV EERDKAAEDS PVNHSQHQCK SNRRNTQRKK 1080 ENLESSNKGN KVVRNRKQFN LETEEEQEGG PSTRLRKRTE KPSKGQGAKS LETKSVAKKQ 1140 PNGLKAKKSP AGSNKMKGKD EKTEYPCDME GCTMGFGSKQ ELVLHKRNIC PVKGCGKKFF 1200 SHKYLVQHRR VHVDERPLKC PWKGCKMSFK WAWARTEHIR VHTGARPYVC AEPGCNQTFR 1260 FVSDFSRHKR KTGHSSSKKG S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 4e-75 | 20 | 376 | 8 | 346 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 4e-75 | 20 | 376 | 8 | 346 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 176 | 182 | GGKEGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
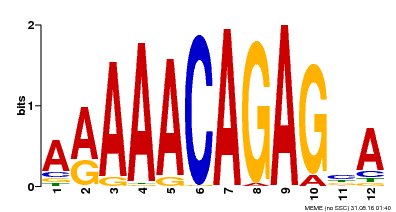
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027123006.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A068V3I4 | 0.0 | A0A068V3I4_COFCA; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400039224 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4988 | 23 | 31 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



