 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc05_g11430 | ||||||||
| Common Name | GSCOC_T00021317001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 354aa MW: 39425.6 Da PI: 7.3188 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 121.7 | 8.5e-38 | 52 | 153 | 2 | 96 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgt.......ssssasec.eaesssssasnsssgkaa 91
+g+kdrhsk++T++g+RdRRvRlsa++a++f+d+qd+LG+d++sk+++WL+++ak ai+el+++ +s +a+ + e++++ +++ +++ ++++
Cc05_g11430 52 TGRKDRHSKVCTAKGPRDRRVRLSAHTAIQFYDVQDRLGYDRPSKAVDWLIKKAKSAIDELAELpawhpttGSAAANTSfEQDQQAQKSRADNLQNQQ 149
79**************************************************************5554433111111111111111111112222222 PP
TCP 92 ksaak 96
+++
Cc05_g11430 150 QQH-L 153
222.2 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 1.9E-31 | 52 | 159 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 34.187 | 54 | 112 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MGENFHHHHQ LQQQQQQQPT TRQSSSRLSG LRNSTGAGEI VEVQGGHIVR STGRKDRHSK 60 VCTAKGPRDR RVRLSAHTAI QFYDVQDRLG YDRPSKAVDW LIKKAKSAID ELAELPAWHP 120 TTGSAAANTS FEQDQQAQKS RADNLQNQQQ QHLGGLHQTD SAANPSGNSS SFLPPSLDSD 180 SIADTIKSFF PMGASAEANS SAMQFQSFPP PDLLSRTSSH PQDLQLSLQS FQDPILLHHH 240 NQQQAQHHQN HPTSQHPEQA EALFSGNTQL GGYDAAAWSE HHQQPAELGR NCSAKTSFFL 300 RGDPFSPLIH LQFVLGWIHQ LYPSPLQIIT FHTIRQCCQF IHHRYLALDL PLE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 1e-20 | 59 | 112 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 1e-20 | 59 | 112 | 1 | 54 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor playing a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164) (PubMed:12931144, PubMed:17307931). Required during early steps of embryogenesis (PubMed:15634699). Participates in ovule develpment (PubMed:25378179). Activates LOX2 expression by binding to the 5'-GGACCA-3' motif found in its promoter (PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:15634699, ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:18816164, ECO:0000269|PubMed:25378179}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00062 | PBM | Transfer from AT3G15030 | Download |
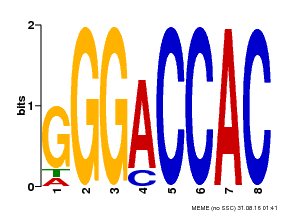
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the miRNA miR-JAW/miR319 (PubMed:12931144, PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:18816164}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027064028.1 | 1e-173 | transcription factor TCP4-like | ||||
| Refseq | XP_027064029.1 | 1e-173 | transcription factor TCP4-like | ||||
| Swissprot | Q8LPR5 | 7e-81 | TCP4_ARATH; Transcription factor TCP4 | ||||
| TrEMBL | A0A068UBV5 | 0.0 | A0A068UBV5_COFCA; Uncharacterized protein | ||||
| STRING | XP_009630327.1 | 1e-104 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3207 | 23 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G15030.3 | 2e-62 | TCP family protein | ||||



