 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc06_g11670 | ||||||||
| Common Name | GSCOC_T00041630001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 281aa MW: 30902.9 Da PI: 8.9764 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 39.3 | 1.5e-12 | 5 | 56 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT eE+e l +vk++G+g Wk I r R+ ++k++w+++
Cc06_g11670 5 KQKWTSEEEEALRAGVKKHGTGKWKNIQRDPEfshllFSRSNIDLKDKWRNL 56
79*********************************9989***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.034 | 1 | 61 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.6E-8 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-14 | 4 | 56 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 5.8E-9 | 5 | 56 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.72E-14 | 6 | 60 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 2.30E-21 | 6 | 57 | No hit | No description |
| SMART | SM00526 | 2.3E-17 | 118 | 183 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 3.54E-15 | 118 | 200 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PROSITE profile | PS51504 | 24.595 | 120 | 188 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene3D | G3DSA:1.10.10.10 | 7.3E-14 | 121 | 188 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00538 | 1.1E-9 | 123 | 182 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 281 aa Download sequence Send to blast |
MGNPKQKWTS EEEEALRAGV KKHGTGKWKN IQRDPEFSHL LFSRSNIDLK DKWRNLTVSA 60 SNGLGPREKS KAQKGKAISD APATPLPITQ VPASSTPLSL EVPADVAADD SSKCLLDGKT 120 ASKYNGMIHE ALSTLKEPNG SDTSTIVNFI EQRHEVPPNF RRLISARLRR LVAQEKLEKV 180 QNSYRIKKDL LEGSKTPVPK PRDVQPRQAQ SIGHLADTVE EATVSAAYTI AEAENKSFVV 240 AEAVKESERV SRMSEDAESM LQLAHEIFDR SSHGEMVLVA * |
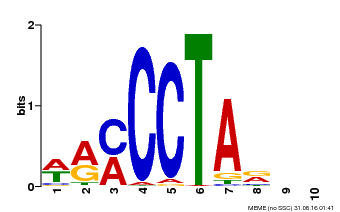
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027068833.1 | 0.0 | telomere repeat-binding factor 4-like | ||||
| TrEMBL | A0A068U3Q4 | 0.0 | A0A068U3Q4_COFCA; Uncharacterized protein | ||||
| STRING | XP_009757029.1 | 1e-129 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2504 | 23 | 55 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72740.2 | 1e-73 | MYB_related family protein | ||||



