 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc06_g17700 | ||||||||
| Common Name | GSCOC_T00040357001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 241aa MW: 27202.1 Da PI: 8.714 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 85.2 | 3.8e-27 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
k+ien rqvtfskRr+g+lKKA+EL +LCdaev vi+fsstgklye++s
Cc06_g17700 9 KKIENVNSRQVTFSKRRAGLLKKAKELAILCDAEVGVIVFSSTGKLYEFAS 59
68***********************************************86 PP
| |||||||
| 2 | K-box | 62.5 | 1.6e-21 | 81 | 170 | 9 | 100 |
K-box 9 leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
e++++++ q +++ Lk+ei +Lq qR++ G++L+ L+++eLq+Le+qL++++ ++++K+++lleq+e++ k +++ en+aLr+++ee
Cc06_g17700 81 MENKTENEPQPQADALKSEIAKLQLVQRQMTGKELGGLNFQELQHLEHQLHEGILAVKDRKEQVLLEQLEKS--KLQKVVLENEALREQVEE 170
455666677899*********************************************************975..667788889999999986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 1.1E-40 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 32.113 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 2.28E-42 | 2 | 77 | No hit | No description |
| SuperFamily | SSF55455 | 2.88E-32 | 2 | 87 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.4E-28 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 9.7E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.4E-28 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.4E-28 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.2E-15 | 82 | 168 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 12.366 | 86 | 174 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010262 | Biological Process | somatic embryogenesis | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048577 | Biological Process | negative regulation of short-day photoperiodism, flowering | ||||
| GO:0060862 | Biological Process | negative regulation of floral organ abscission | ||||
| GO:0060867 | Biological Process | fruit abscission | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 241 aa Download sequence Send to blast |
MGRGKIEIKK IENVNSRQVT FSKRRAGLLK KAKELAILCD AEVGVIVFSS TGKLYEFAST 60 RMEQILARYN TNPESSKLIT MENKTENEPQ PQADALKSEI AKLQLVQRQM TGKELGGLNF 120 QELQHLEHQL HEGILAVKDR KEQVLLEQLE KSKLQKVVLE NEALREQVEE YRLRLYRENG 180 QLGRKNSGVC SSTVCDCRSE KDGNSDTSLR LGLSVDICHK RKKPKTESTS NDSETLTVLE 240 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 6e-22 | 1 | 88 | 1 | 94 | MEF2C |
| 5f28_B | 6e-22 | 1 | 88 | 1 | 94 | MEF2C |
| 5f28_C | 6e-22 | 1 | 88 | 1 | 94 | MEF2C |
| 5f28_D | 6e-22 | 1 | 88 | 1 | 94 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. | |||||
| UniProt | Probable transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Prevents premature flowering. Downstream regulator of a subset of the MIKC* MADS-controlled genes required during pollen maturation. {ECO:0000269|PubMed:17521410, ECO:0000269|PubMed:18034896, ECO:0000269|PubMed:18799658}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00508 | DAP | Transfer from AT5G13790 | Download |
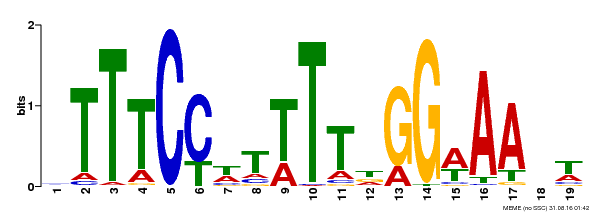
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027069013.1 | 1e-175 | MADS-box transcription factor 23-like isoform X2 | ||||
| Swissprot | Q39295 | 9e-59 | AGL15_BRANA; Agamous-like MADS-box protein AGL15 | ||||
| Swissprot | Q9M2K8 | 8e-59 | AGL18_ARATH; Agamous-like MADS-box protein AGL18 | ||||
| TrEMBL | A0A068V010 | 1e-173 | A0A068V010_COFCA; Uncharacterized protein | ||||
| STRING | XP_009758657.1 | 4e-89 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G57390.1 | 3e-46 | AGAMOUS-like 18 | ||||



