 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc07_g02340 | ||||||||
| Common Name | GSCOC_T00039421001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 280aa MW: 31618.9 Da PI: 4.417 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.1 | 9.5e-21 | 64 | 117 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t+eq++ Le+ Fe ++++ e++++LAkklgL+ rqV vWFqNrRa++k
Cc07_g02340 64 KKRRLTPEQVHLLEKSFEAENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWK 117
56689************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.6 | 1.3e-41 | 63 | 153 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ eqv+lLE+sFe+e+kLeperK++la++LglqprqvavWFqnrRAR+ktkqlE+dy+++k++yd+l+++ +++ ke+e+L++e+
Cc07_g02340 63 EKKRRLTPEQVHLLEKSFEAENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWKTKQLERDYDQVKSSYDSLRSDYDSVVKENEKLKTEV 153
69*************************************************************************************9887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 17.993 | 59 | 119 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 8.0E-20 | 62 | 123 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.11E-19 | 64 | 129 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 5.8E-18 | 64 | 117 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.26E-19 | 64 | 120 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-22 | 65 | 126 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.2E-6 | 90 | 99 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 94 | 117 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.2E-6 | 99 | 115 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 3.1E-17 | 119 | 161 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 280 aa Download sequence Send to blast |
MDSGRIYFDP SCHGNMLFLG SGNPVFPGAR SVLNVEETLK RRPFLRSPEE LFDEEYYEEQ 60 LAEKKRRLTP EQVHLLEKSF EAENKLEPER KTQLAKKLGL QPRQVAVWFQ NRRARWKTKQ 120 LERDYDQVKS SYDSLRSDYD SVVKENEKLK TEVLSLTEKL QQGKEVVVEP RSKQKSDALP 180 VDELLASDPQ FNVKVEDRLS TGSGGSAVVD EDGPQLVDSG DSYFPCDDYA AGCVTPAADG 240 VQSEEDDGSD NGQNYLSNVF VAAEQQNQEG EPVGWWVWS* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 111 | 119 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
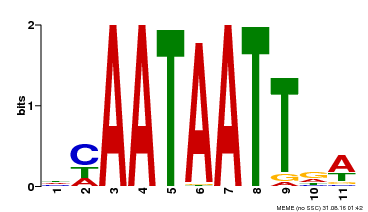
| UniProt | Probable transcription activator involved in leaf development. Binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:8535134}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027074934.1 | 0.0 | homeobox-leucine zipper protein HAT5-like isoform X1 | ||||
| Swissprot | Q02283 | 3e-95 | HAT5_ARATH; Homeobox-leucine zipper protein HAT5 | ||||
| TrEMBL | A0A068U1T9 | 0.0 | A0A068U1T9_COFCA; Uncharacterized protein | ||||
| STRING | EOY02359 | 1e-140 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA7891 | 23 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 6e-89 | homeobox 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



