 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cc08_g11910 | ||||||||
| Common Name | GSCOC_T00030556001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Rubiaceae; Ixoroideae; Coffeeae; Coffea
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 228aa MW: 24869.7 Da PI: 9.3199 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.2 | 1.1e-19 | 88 | 139 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkklege 56
+y+GVr+++ +g+++AeIrdp +ng +r++lg+f tae+Aa a+++a+ +++g+
Cc08_g11910 88 HYRGVRRRP-WGKFAAEIRDPAKNG--ARVWLGTFETAEDAAIAYDRAAYRMRGA 139
8********.***********9987..**************************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 9.8E-32 | 87 | 146 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.38E-29 | 87 | 146 | No hit | No description |
| SuperFamily | SSF54171 | 2.22E-23 | 88 | 148 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 24.395 | 88 | 146 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.3E-36 | 88 | 152 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.4E-13 | 88 | 138 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.2E-11 | 89 | 100 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.2E-11 | 112 | 128 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0009864 | Biological Process | induced systemic resistance, jasmonic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 228 aa Download sequence Send to blast |
MTYGNGDLQA DLALLESIRQ HLLEESEASA PALCFYNSRP VSYSGSNPSE ASFFPSFPDH 60 EERIEVSQMS NAATAQPEAR VVPPKGKHYR GVRRRPWGKF AAEIRDPAKN GARVWLGTFE 120 TAEDAAIAYD RAAYRMRGAR ALLNFPLRIN SGEPEPVRIT SKRSSVSPDH SSSSASENGT 180 PKRKKKAAAQ VVEPPAVQRG WEELYVGSNS NGERIFATGT QFGSTLL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 4e-42 | 85 | 153 | 2 | 70 | ATERF1 |
| 3gcc_A | 4e-42 | 85 | 153 | 2 | 70 | ATERF1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. Involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. Involved in disease resistance pathways. {ECO:0000269|PubMed:10715325, ECO:0000269|PubMed:12805630, ECO:0000269|PubMed:9756931}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00548 | DAP | Transfer from AT5G47220 | Download |
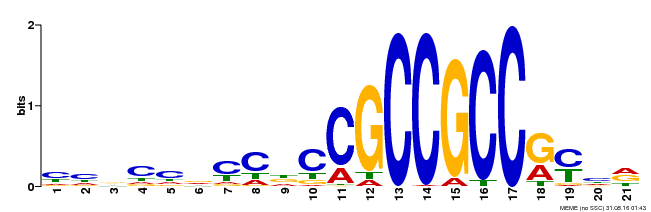
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by jasmonate (JA) and Alternaria brassicicola (locally and systemically). Ethylene induction is completely dependent on a functional ETHYLENE-INSENSITIVE2 (EIN2) while wounding induction does not require EIN2. Transcripts accumulate strongly in cycloheximide-treated plants, a protein synthesis inhibitor. Seems to not be influenced by exogenous abscisic acid (ABA), cold, heat, NaCl or drought stress. {ECO:0000269|PubMed:10715325, ECO:0000269|PubMed:12805630}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027085492.1 | 1e-139 | ethylene-responsive transcription factor 2-like | ||||
| Swissprot | O80338 | 3e-54 | EF101_ARATH; Ethylene-responsive transcription factor 2 | ||||
| TrEMBL | A0A068UQX4 | 1e-165 | A0A068UQX4_COFCA; Uncharacterized protein | ||||
| STRING | GLYMA17G15480.1 | 5e-57 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA21 | 24 | 1165 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47220.1 | 7e-42 | ethylene responsive element binding factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



