 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ciclev10005102m | ||||||||
| Common Name | CICLE_v10005102mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 402aa MW: 44840.7 Da PI: 4.8349 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.2 | 2.5e-15 | 28 | 75 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+W++ Ed++l+ ++++G ++W++ ++ g+ R++k+c++rw +yl
Ciclev10005102m 28 RGPWSPGEDLRLITFIQKHGHENWRALPKQAGLLRCGKSCRLRWINYL 75
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 52.4 | 1.3e-16 | 81 | 126 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+eE++ ++++++ lG++ W++Ia+ ++ gRt++++k+ w+++l
Ciclev10005102m 81 RGNFTQEEEDTIIRLHESLGNR-WSKIASQLP-GRTDNEIKNVWNTHL 126
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-23 | 19 | 78 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.096 | 23 | 75 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.87E-29 | 26 | 122 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.0E-12 | 27 | 77 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.2E-14 | 28 | 75 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.36E-8 | 30 | 75 | No hit | No description |
| PROSITE profile | PS51294 | 25.592 | 76 | 130 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.1E-27 | 79 | 130 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-15 | 80 | 128 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-15 | 81 | 126 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.19E-10 | 83 | 126 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 402 aa Download sequence Send to blast |
MVGSLKIFLL SQMGKGRAPC CDKSQVKRGP WSPGEDLRLI TFIQKHGHEN WRALPKQAGL 60 LRCGKSCRLR WINYLRPDVK RGNFTQEEED TIIRLHESLG NRWSKIASQL PGRTDNEIKN 120 VWNTHLKKRL SFRDVKKDQE SKESSSISSP SSSSSSTIIS SGKRRADTEL DEHQCDQNNY 180 VPKKPREGHG SMTVSEKLVQ QSSETEESNQ INTSKGVVMT TKELLDSSCS SDNSVVSNSS 240 QVDATRPKDQ MGTSQFGFSE PYDVAIGLNN LEEVNKPEII SISDTGLEIP LECDFDFWSM 300 LDNLGPFQSH EVEASQSTSF GEASDNSREV DSRTWFQYLE NELGLEATTE DEIQNSAKVA 360 AAATADPIPQ ETYETLLKPE VDPGVTYFQL WNASLQSSVC E* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 4e-27 | 28 | 130 | 4 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 4e-27 | 28 | 130 | 4 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
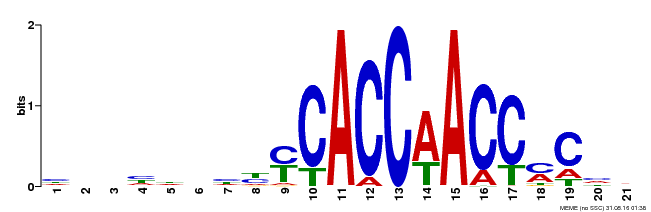
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00249 | DAP | Transfer from AT1G79180 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006475333.1 | 0.0 | transcription factor MYB63 | ||||
| Refseq | XP_024038620.1 | 0.0 | transcription factor MYB63 | ||||
| TrEMBL | V4RMI1 | 0.0 | V4RMI1_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006422238.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4 | 28 | 2646 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G16490.1 | 4e-76 | myb domain protein 58 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Ciclev10005102m |
| Entrez Gene | 18031549 |



