 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ciclev10008110m | ||||||||
| Common Name | CICLE_v10007904mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 493aa MW: 52971.8 Da PI: 4.5086 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.8 | 1.9e-32 | 302 | 359 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkkkvers +++++ei+Y+g Hnh+k
Ciclev10008110m 302 EDGYNWRKYGQKQVKGSEYPRSYYKCTHPNCQVKKKVERSL-EGHITEIIYKGAHNHPK 359
7****************************************.***************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 22.848 | 296 | 360 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.2E-28 | 300 | 360 | IPR003657 | WRKY domain |
| SMART | SM00774 | 6.8E-36 | 301 | 359 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.57E-25 | 301 | 360 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 9.6E-25 | 302 | 358 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 493 aa Download sequence Send to blast |
MVMAGIDDNM AVIGDWVPPS PSPRTFFAAI SGDDISSREP PSENGVDGLS LGSQEQMTSG 60 NTEKKNMMTG GASTDLGPFS EQKSRLAARA GFNAPRLNTE SIRTSDLSLN PDIRSPYLTI 120 PPGLSPTTLL ESPVFLSNSL AQPSPTTGKF SFIPNGNGKS STCISEAPDK SKDNILEDIN 180 TSFAFKPVAE SGSFSLGTRN KQSFPSIEVS VQSENSPQNV NAAKVHSQYR NSLSLQVQTD 240 FSRATTEKDN GCNTILADQR VFDTFGGSTE HSPPLDEQQD EEGDQRGSGD SMVAGAGGAS 300 SEDGYNWRKY GQKQVKGSEY PRSYYKCTHP NCQVKKKVER SLEGHITEII YKGAHNHPKP 360 PPNRRSAIGS NPLDMQLDIH EQAGLQSGAD GDPSWANTQK GTAGGTPEWR HDTVEVTSAA 420 PVGPEYCNQS SALPAQNGTP YESGDAVDAS STFSNDEDED DQVTHCSVGF DGEEDESESK 480 RRYLFPFSIF LL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 1e-17 | 303 | 361 | 16 | 75 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Low expression in senescent leaves (PubMed:11722756). Expressed in both the unfertilized egg cell and the pollen tube (PubMed:21316593). {ECO:0000269|PubMed:11722756, ECO:0000269|PubMed:21316593}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
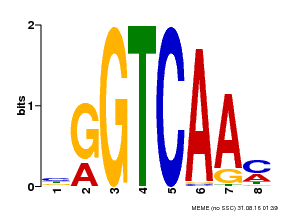
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024033318.1 | 0.0 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_024033319.1 | 0.0 | probable WRKY transcription factor 2 isoform X1 | ||||
| Swissprot | Q9FG77 | 1e-107 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | V4UIP7 | 0.0 | V4UIP7_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006474751.1 | 0.0 | (Citrus sinensis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56270.1 | 1e-110 | WRKY DNA-binding protein 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Ciclev10008110m |
| Entrez Gene | 18054799 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



