 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ciclev10011446m | ||||||||
| Common Name | CICLE_v10011452mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 536aa MW: 58929 Da PI: 8.8294 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.1 | 7.5e-17 | 60 | 106 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde l +av + g++Wk+Ia+ ++ R++ qc +rwqk+l
Ciclev10011446m 60 KGGWTPEEDETLRNAVSTFKGKSWKKIAEFFP-DRSEVQCLHRWQKVL 106
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 59.1 | 1e-18 | 112 | 158 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+++ ++v ++G+ W+ Ia+ ++ gR +kqc++rw+++l
Ciclev10011446m 112 KGPWTQEEDDKITELVSKYGPTKWSVIAKSLP-GRIGKQCRERWHNHL 158
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 56 | 8.8e-18 | 164 | 207 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WT eE++ l +a++ +G++ W+ Ia+ ++ gRt++++k++w++
Ciclev10011446m 164 KDAWTLEEELALMNAHRIHGNK-WAEIAKVLP-GRTDNSIKNHWNS 207
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.551 | 55 | 106 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.2E-14 | 59 | 108 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.5E-15 | 60 | 106 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.94E-15 | 61 | 116 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.7E-20 | 62 | 115 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.80E-13 | 63 | 106 | No hit | No description |
| PROSITE profile | PS51294 | 31.365 | 107 | 162 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.25E-31 | 109 | 205 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.0E-17 | 111 | 160 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.1E-18 | 112 | 158 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.58E-15 | 114 | 158 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-27 | 116 | 165 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-16 | 163 | 211 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 22.781 | 163 | 213 | IPR017930 | Myb domain |
| Pfam | PF00249 | 5.9E-16 | 164 | 207 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-23 | 166 | 213 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 8.46E-13 | 166 | 209 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 536 aa Download sequence Send to blast |
MAEVKLEEEC CLENKQLTAA SSSSLSEGSS SAILKSPGVS SPATTSPTHR RTTGPIRRAK 60 GGWTPEEDET LRNAVSTFKG KSWKKIAEFF PDRSEVQCLH RWQKVLNPDL VKGPWTQEED 120 DKITELVSKY GPTKWSVIAK SLPGRIGKQC RERWHNHLNP DIKKDAWTLE EELALMNAHR 180 IHGNKWAEIA KVLPGRTDNS IKNHWNSSLK KKLDFYLATG KLPPVAKSSL QNGTKDTSQS 240 TATENLVCSN KDSDSAAQTS SGTTDIGKPD EEGGKDRLEP SALVPDMATS SSIRPNESMD 300 SEGLECNPES PNIDLTCSES MPRLENCTVN CEFVEDKVTG TQKQVGTPTY GSLYYVPPEL 360 KSFILLDKDT SNRHSVHHDY NSSPITSPIS FFTPPCVKGS GLSSPSPESI LKIAAKTFPY 420 TPSIFRKRKP VSQAQLDAKK IGKVDGVSGS DRFCLSGKQE SNQNNSKNSR FQDASPSGSI 480 EPTGMTFNAS PPYRLRSKRT AVFKSVERQL EFTSNKEKGD RNANFVKQSV KGTSH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 6e-68 | 59 | 213 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 6e-68 | 59 | 213 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed both in proliferating and maturing stages of leaves. {ECO:0000269|PubMed:26069325}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
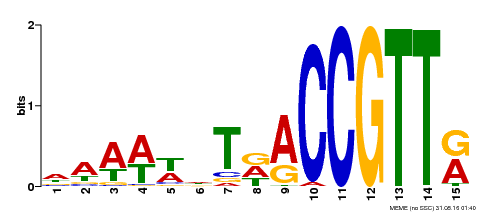
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006429084.1 | 0.0 | transcription factor MYB3R-5 | ||||
| Refseq | XP_006429085.1 | 0.0 | transcription factor MYB3R-5 | ||||
| Refseq | XP_006480818.1 | 0.0 | transcription factor MYB3R-5 | ||||
| Refseq | XP_006480819.1 | 0.0 | transcription factor MYB3R-5 | ||||
| Refseq | XP_006480820.1 | 0.0 | transcription factor MYB3R-5 | ||||
| Refseq | XP_024037722.1 | 0.0 | transcription factor MYB3R-5 | ||||
| Swissprot | Q6R032 | 1e-175 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | A0A2H5NYB1 | 0.0 | A0A2H5NYB1_CITUN; Uncharacterized protein | ||||
| TrEMBL | V4US04 | 0.0 | V4US04_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006480818.1 | 0.0 | (Citrus sinensis) | ||||
| STRING | XP_006429084.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1667 | 28 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-166 | myb domain protein 3r-5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Ciclev10011446m |
| Entrez Gene | 18037936 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



