 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ciclev10011554m | ||||||||
| Common Name | CICLE_v10011554mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 497aa MW: 54586.8 Da PI: 7.4156 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.8 | 1.9e-16 | 60 | 104 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE++++++a k++G W++I +++g ++t+ q++s+ qk+
Ciclev10011554m 60 RERWTEEEHKKFLEALKLFGRA-WRKIEEHVG-TKTAVQIRSHAQKF 104
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 9.72E-17 | 54 | 108 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.193 | 55 | 109 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 8.2E-17 | 58 | 107 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 3.7E-13 | 59 | 107 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-9 | 60 | 100 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.8E-14 | 60 | 103 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.49E-10 | 62 | 105 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 497 aa Download sequence Send to blast |
MAVQDECGGN HSNSVFSAAN GIESSSGLHT VTGLEVKDQF SCGNDFAPKP RKPYTITKQR 60 ERWTEEEHKK FLEALKLFGR AWRKIEEHVG TKTAVQIRSH AQKFFSKVVR ESNGCSTSPV 120 EPVEIPPPRP KRKPMHPYPR KLAHPPVKES LNPELSRTSL SPILSVSERE NQSPTSVLFA 180 IGSDAFGSSD SNSPNGSLSP VSSAVPEQLG GLTLSHPSSS PEERGSPSSD PVTPGSVTDE 240 QSPKCVLKMG LSPQRFDKEV SDVRASSRSL KLFGTIVLVT DSQRLSSPTA GTCKLFESFP 300 SEALEGKPLQ LLPLEVMHTE SLSEDAKYAW RRTLPHGGSG AFSCMQSTDD DSNMVDTGSG 360 AHLPWWNFSH GLPYAVTPNN QQVQNLRLHS SVKEVQGREF KKEGSLTGSN SKSVNEEDSI 420 DKSSDVDSQS SRQSLDKEDK WPEVAFELKP SENSAFSVIR TRTDKHMKGF VPYKKRIVER 480 DNQLSAVGNG RDQREW* |
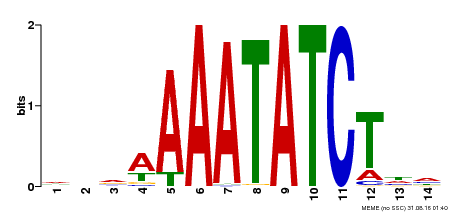
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006430409.1 | 0.0 | protein REVEILLE 1 | ||||
| TrEMBL | V4UVE9 | 0.0 | V4UVE9_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006430409.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6356 | 26 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 4e-53 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Ciclev10011554m |
| Entrez Gene | 18039807 |



